Loading AI tools
Type of paternal lineage From Wikipedia, the free encyclopedia
Haplogroup R1b (R-M343), previously known as Hg1 and Eu18, is a human Y-chromosome haplogroup.
| Haplogroup R1b | |
|---|---|
 | |
| Possible time of origin | Probably soon after R1, possibly between 18,000-14,000 BC[1] |
| Possible place of origin | Western Asia, North Eurasia or Eastern Europe[2] |
| Ancestor | R1 |
| Descendants |
|
| Defining mutations | M343 |
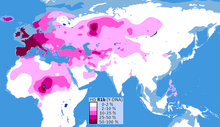
It is the most frequently occurring paternal lineage in Western Europe, as well as some parts of Russia (e.g. the Bashkirs) and across the Sahel in Central Africa, namely: Cameroon, Chad, Guinea, Mauritania, Mali, Niger, Nigeria and Senegal (concentrated in parts of Chad with concentration in the Hausa Tribe and among the Chadic-speaking ethnic groups of Cameroon).
The clade is also present at lower frequencies throughout Eastern Europe, Western Asia, Central Asia as well as parts of North Africa, South Asia and Central Asia.
R1b has two primary branches: R1b1-L754 and R1b2-PH155. R1b1-L754 has two major subclades: R1b1a1b-M269, which predominates in Western Europe, and R1b1b-V88, which is today common in parts of Central Africa. The other branch, R1b2-PH155, is so rare and widely dispersed that it is difficult to draw any conclusions about its origins. It has been found in Bahrain, India, Nepal, Bhutan, Ladakh, Tajikistan, Turkey, and Western China.
According to ancient DNA studies, most R1a and R1b lineages would have expanded from the Pontic Steppe along with the Indo-European languages.[2][3][4][5][6]
The age of R1 was estimated by Tatiana Karafet et al. (2008) at between 12,500 and 25,700 BP, and most probably occurred about 18,500 years ago.[7] Since the earliest known example has been dated at circa 14,000 BP, and belongs to R1b1 (R-L754),[1] R1b must have arisen relatively soon after the emergence of R1.
Early human remains found to carry R1b include:
R1b is a subclade within the "macro-haplogroup" K (M9), the most common group of human male lines outside of Africa. K is believed to have originated in Asia (as is the case with an even earlier ancestral haplogroup, F (F-M89). Karafet T. et al. (2014) suggested that a "rapid diversification process of K-M526 likely occurred in Southeast Asia, with subsequent westward expansions of the ancestors of haplogroups R and Q".[29] However the oldest example of R* has been found in an Ancient North Eurasian sample from Siberia (Mal'ta boy, 24,000 years ago), and its precursor P1 has been found in another Ancient North Eurasian sample from northern Siberia (Yana RHS) dating from c. 31,600 years ago.[30]
Three genetic studies in 2015 gave support to the Kurgan hypothesis of Marija Gimbutas regarding the Proto-Indo-European homeland. According to those studies, haplogroups R1b-M269 and R1a, now the most common in Europe (R1a is also common in South Asia) would have expanded from the West Eurasian Steppe, along with the Indo-European languages; they also detected an autosomal component present in modern Europeans which was not present in Neolithic Europeans, which would have been introduced with paternal lineages R1b and R1a, as well as Indo-European languages.[2][3][4]

Analysis of ancient Y-DNA from the remains from early Neolithic Central and North European Linear Pottery culture settlements have not yet found males belonging to haplogroup R1b-M269.[31][32] Olalde et al. (2017) trace the spread of haplogroup R1b-M269 in western Europe, particularly Britain, to the spread of the Beaker culture, with a sudden appearance of many R1b-M269 haplogroups in Western Europe ca. 5000–4500 years BP during the early Bronze Age.[33]
The broader haplogroup R (M207) is a primary subclade of haplogroup P1 (M45) itself a primary branch of P (P295), which is also known as haplogroup K2b2. R-M207 is therefore a secondary branch of K2b (P331), and a direct descendant of K2 (M526).
Names such as R1b, R1b1 and so on are phylogenetic (i.e. "family tree") names which make clear their place within the branching of haplogroups, or the phylogenetic tree. An alternative way of naming the same haplogroups and subclades refers to their defining SNP mutations: for example, R-M343 is equivalent to R1b.[34] Phylogenetic names change with new discoveries and SNP-based names are consequently reclassified within the phylogenetic tree. In some cases, an SNP is found to be unreliable as a defining mutation and an SNP-based name is removed completely. For example, before 2005, R1b was synonymous with R-P25, which was later reclassified as R1b1; in 2016, R-P25 was removed completely as a defining SNP, due to a significant rate of back-mutation.[35] (Below is the basic outline of R1b according to the ISOGG Tree as it stood on January 30, 2017.[36])
|
No confirmed cases of R1b* (R-M343*) – that is R1b (xR1b1, R1b2), also known as R-M343 (xL754, PH155) – have been reported in peer-reviewed literature.
In early research, because R-M269, R-M73 and R-V88 are by far the most common forms of R1b, examples of R1b (xM73, xM269) were sometimes assumed to signify basal examples of "R1b*".[35] However, while the paragroup R-M343 (xM73, M269, V88) is rare, it does not preclude membership of rare and/or subsequently-discovered, relatively basal subclades of R1b, such as R-L278* (R1b*), R-L389* (R1b1a*), R-P297* (R1b1a1*), R-V1636 (R1b1a2) or R-PH155 (R1b2).
The population believed to have the highest proportion of R-M343 (xM73, M269, V88) are the Kurds of southeastern Kazakhstan with 13%.[41] However, more recently, a large study of Y-chromosome variation in Iran, revealed R-M343 (xV88, M73, M269) as high as 4.3% among Iranian sub-populations.[42]
It remains a possibility that some, or even most of these cases, may be R-L278* (R1b*), R-L389* (R1b1a*), R-P297* (R1b1a1*), R-V1636 (R1b1a2), R-PH155 (R1b2), R1b* (R-M343*), R1a* (R-M420*), an otherwise undocumented branch of R1 (R-M173), and/or back-mutations of a marker, from a positive to a negative ancestral state,[43] and hence constitute undocumented subclades of R1b.
A compilation of previous studies regarding the distribution of R1b can be found in Cruciani et al. (2010).[44] It is summarised in the table following. (Cruciani did not include some studies suggesting even higher frequencies of R1b1a1b [R-M269] in some parts of Western Europe.)
| Continent | Region | Sample size |
Total R1b | R-P25 (unreliable marker for R1b1*) |
R-V88 (R1b1b) |
R-M269 (R1b1a1a2) |
R-M73 (R1b1a1a1) |
|---|---|---|---|---|---|---|---|
| Africa | Northern Africa | 691 | 5.9% | 0.0% | 5.2% | 0.7% | 0.0% |
| Africa | Central Sahel Region | 461 | 23.0% | 0.0% | 23.0% | 0.0% | 0.0% |
| Africa | Western Africa | 123 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Africa | Eastern Africa | 442 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Africa | Southern Africa | 105 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Europe | Western Europeans | 465 | 57.8% | 0.0% | 0.0% | 57.8% | 0.0% |
| Europe | North-west Europeans | 43 | 55.8% | 0.0% | 0.0% | 55.8% | 0.0% |
| Europe | Central Europeans | 77 | 42.9% | 0.0% | 0.0% | 42.9% | 0.0% |
| Europe | North Eastern Europeans | 74 | 1.4% | 0.0% | 0.0% | 1.4% | 0.0% |
| Europe | Russians | 60 | 6.7% | 0.0% | 0.0% | 6.7% | 0.0% |
| Europe | Eastern Europeans | 149 | 20.8% | 0.0% | 0.0% | 20.8% | 0.0% |
| Europe | South-east Europeans | 510 | 13.1% | 0.0% | 0.2% | 12.9% | 0.0% |
| Asia | West Asians | 328 | 5.8% | 0.0% | 0.3% | 5.5% | 0.0% |
| Asia | South Asians | 288 | 4.8% | 0.0% | 0.0% | 1.7% | 3.1% |
| Asia | South-east Asians | 10 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Asia | North-east Asians | 30 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Asia | East Asians | 156 | 0.6% | 0.0% | 0.0% | 0.6% | 0.0% |
| Total | 5326 | ||||||
R-L278 among modern men falls into the R-L754 and R-PH155 subclades, though it is possible some very rare R-L278* may exist as not all examples have been tested for both branches. Examples may also exist in ancient DNA, though due to poor quality it is often impossible to tell whether or not the ancients carried the mutations that define subclades.
Some examples described in older articles, for example two found in Turkey,[34] are now thought to be mostly in the more recently discovered sub-clade R1b1b (R-V88). Most examples of R1b therefore fall into subclades R1b1b (R-V88) or R1b1a (R-P297). Cruciani et al. in the large 2010 study found 3 cases amongst 1173 Italians, 1 out of 328 West Asians and 1 out of 156 East Asians.[44] Varzari found 3 cases in Ukraine, in a study of 322 people from the Dniester–Carpathian Mountains region, who were P25 positive, but M269 negative.[45] Cases from older studies are mainly from Africa, the Middle East or Mediterranean, and are discussed below as probable cases of R1b1b (R-V88).
R-L754 contains the vast majority of R1b. The only known example of R-L754* (xL389, V88) is also the earliest known individual to carry R1b: "Villabruna 1", who lived circa 14,000 years BP (north east Italy). Villabruna 1 belonged to the Epigravettian culture.
R-L389, also known as R1b1a (L388/PF6468, L389/PF6531), contains the very common subclade R-P297 and the rare subclade R-V1636. It is unknown whether all previously reported R-L389* (xP297) belong to R-V1636 or not.
The SNP marker P297 was recognised in 2008 as ancestral to the significant subclades M73 and M269, combining them into one cluster.[7] This had been given the phylogenetic name R1b1a1a (and, previously, R1b1a).
A majority of Eurasian R1b falls within this subclade, representing a very large modern population. Although P297 itself has not yet been much tested for, the same population has been relatively well studied in terms of other markers. Therefore, the branching within this clade can be explained in relatively high detail below.
Malyarchuk et al. (2011) found R-M73 in 13.2% (5/38) of Shors, 11.4% (5/44) of Teleuts, 3.3% (2/60) of Kalmyks, 3.1% (2/64) of Khakassians, 1.9% (2/108) of Tuvinians, and 1.1% (1/89) of Altaians.[46] The Kalmyks, Tuvinians, and Altaian belong to a Y-STR cluster marked by DYS390=19, DYS389=14-16 (or 14–15 in the case of the Altaian individual), and DYS385=13-13.
Dulik et al. (2012) found R-M73 in 35.3% (6/17) of a sample of the Kumandin of the Altai Republic in Russia.[47] Three of these six Kumandins share an identical 15-loci Y-STR haplotype, and another two differ only at the DYS458 locus, having DYS458=18 instead of DYS458=17. This pair of Kumandin R-M73 haplotypes resembles the haplotypes of two Kalmyks, two Tuvinians, and one Altaian whose Y-DNA has been analyzed by Malyarchuk et al. (2011). The remaining R-M73 Kumandin has a Y-STR haplotype that is starkly different from the haplotypes of the other R-M73 Kumandins, resembling instead the haplotypes of five Shors, five Teleuts, and two Khakassians.[46]
While early research into R-M73 claimed that it was significantly represented among the Hazara of Afghanistan and the Bashkirs of the Ural Mountains, this has apparently been overturned. For example, supporting material from a 2010 study by Behar et al. suggested that Sengupta et al. (2006) might have misidentified Hazara individuals, who instead belonged to "PQR2" as opposed to "R(xR1a)."[48][41][49] However, the assignment of these Hazaras' Y-DNA to the "PQR2" category by Behar et al. (2010) is probably ascribable to the habit that was popular for a while of labeling R-M269 as "R1b" or "R(xR1a)," with any members of R-M343 (xM269) being placed in a polyphyletic, catch-all "R*" or "P" category. Myres et al. (2011), Di Cristofaro et al. (2013), and Lippold et al. (2014) all agree that the Y-DNA of 32% (8/25) of the HGDP sample of Pakistani Hazara should belong to haplogroup R-M478/M73.[41][50][51] Likewise, most Bashkir males have been found to belong to U-152 (R1b1a1a2a1a2b) and some, mostly from southeastern Bashkortostan, belonged to Haplogroup Q-M25 (Q1a1b) rather than R1b; contra this, Myres et al. (2011) found a high frequency of R-M73 among their sample of Bashkirs from southeast Bashkortostan (77/329 = 23.4% R1b-M73), in agreement with the earlier study of Bashkirs.[41] Besides the high frequency of R-M73 in southeastern Bashkirs, Myres et al. also reported finding R-M73 in the following samples: 10.3% (14/136) of Balkars from the northwest Caucasus, 9.4% (8/85) of the HGDP samples from northern Pakistan (these are the aforementioned Pakistani Hazaras), 5.8% (4/69) of Karachays from the northwest Caucasus, 2.6% (1/39) of Tatars from Bashkortostan, 1.9% (1/54) of Bashkirs from southwest Bashkortostan, 1.5% (1/67) of Megrels from the south Caucasus, 1.4% (1/70) of Bashkirs from north Bashkortostan, 1.3% (1/80) of Tatars from Kazan, 1.1% (1/89) of a sample from Cappadocia, Turkey, 0.7% (1/141) of Kabardians from the northwest Caucasus, 0.6% (3/522) of a pool of samples from Turkey, and 0.38% (1/263) of Russians from Central Russia.[41]
Besides the aforementioned Pakistani Hazaras, Di Cristofaro et al. (2013) found R-M478/M73 in 11.1% (2/18) of Mongols from central Mongolia, 5.0% (1/20) of Kyrgyz from southwest Kyrgyzstan, 4.3% (1/23) of Mongols from southeast Mongolia, 4.3% (4/94) of Uzbeks from Jawzjan, Afghanistan, 3.7% (1/27) of Iranians from Gilan, 2.5% (1/40) of Kyrgyz from central Kyrgyzstan, 2.1% (2/97) of Mongols from northwest Mongolia, and 1.4% (1/74) of Turkmens from Jawzjan, Afghanistan.[50] The Mongols as well as the individual from southwest Kyrgyzstan, the individual from Gilan, and one of the Uzbeks from Jawzjan belong to the same Y-STR haplotype cluster as five of six Kumandin members of R-M73 studied by Dulik et al. (2012). This cluster's most distinctive Y-STR value is DYS390=19.[41]
Karafet et al. (2018) found R-M73 in 37.5% (15/40) of a sample of Teleuts from Bekovo, Kemerovo oblast, 4.5% (3/66) of a sample of Uyghurs from Xinjiang Uyghur Autonomous Region, 3.4% (1/29) of a sample of Kazakhs from Kazakhstan, 2.3% (3/129) of a sample of Selkups, 2.3% (1/44) of a sample of Turkmens from Turkmenistan, and 0.7% (1/136) of a sample of Iranians from Iran.[52] Four of these individuals (one of the Teleuts, one of the Uyghurs, the Kazakh, and the Iranian) appear to belong to the aforementioned cluster marked by DYS390=19 (the Kumandin-Mongol R-M73 cluster); the Teleut and the Uyghur also share the modal values at the DYS385 and the DYS389 loci. The Iranian differs from the modal for this cluster by having 13-16 (or 13–29) at DYS389 instead of 14-16 (or 14–30). The Kazakh differs from the modal by having 13–14 at DYS385 instead of 13-13. The other fourteen Teleuts and the three Selkups appear to belong to the Teleut-Shor-Khakassian R-M73 cluster from the data set of Malyarchuk et al. (2011); this cluster has the modal values of DYS390=22 (but 21 in the case of two Teleuts and one Khakassian), DYS385=13-16, and DYS389=13-17 (or 13–30, but 14–31 in the case of one Selkup).
A Kazakhstani paper published in 2017 found haplogroup R1b-M478 Y-DNA in 3.17% (41/1294) of a sample of Kazakhs from Kazakhstan, with this haplogroup being observed with greater than average frequency among members of the Qypshaq (12/29 = 41.4%), Ysty (6/57 = 10.5%), Qongyrat (8/95 = 8.4%), Oshaqty (2/29 = 6.9%), Kerey (1/28 = 3.6%), and Jetyru (3/86 = 3.5%) tribes.[53] A Chinese paper published in 2018 found haplogroup R1b-M478 Y-DNA in 9.2% (7/76) of a sample of Dolan Uyghurs from Horiqol township, Awat County, Xinjiang.[54]
R-M269, or R1b1a1b (as of 2018) amongst other names,[55] is now the most common Y-DNA lineage in European males. It is carried by an estimated 110 million males in Europe.[56]

R-M269 has received significant scientific and popular interest due to its possible connection to the Indo-European expansion in Europe. Specifically the R-Z2103 subclade has been found to be prevalent in ancient DNA associated with the Yamna culture.[2] All seven individuals in one were determined to belong to the R1b-M269 subclade.[2]
Older research, published before researchers could study the DNA of ancient remains, proposed that R-M269 likely originated in Western Asia and was present in Europe by the Neolithic period.[36][41][57][58] But results based on actual ancient DNA noticed that there was a dearth of R-M269 in Europe before the Bronze Age,[2] and the distribution of subclades within Europe is substantially due to the various migrations of the Bronze and Iron Age. Likewise, the oldest samples classified as belonging to R-M269, have been found in Eastern Europe and Pontic-Caspian steppe, not Western Asia. Western European populations are divided between the R-P312/S116 and R-U106/S21 subclades of R-M412 (R-L51).
Distribution of R-M269 in Europe increases in frequency from east to west. It peaks at the national level in Wales at a rate of 92%, at 82% in Ireland, 70% in Scotland, 68% in Spain, 60% in France (76% in Normandy), about 60% in Portugal,[41] 50% in Germany, 50% in the Netherlands, 47% in Italy,[59] 45% in Eastern England and 42% in Iceland. R-M269 reaches levels as high as 95% in parts of Ireland. It has also been found at lower frequencies throughout central Eurasia,[60] but with relatively high frequency among the Bashkirs of the Perm region (84.0%).[61] This marker is present in China and India at frequencies of less than one percent. In North Africa and adjoining islands, while R-V88 (R1b1b) is more strongly represented, R-M269 appears to have been present since antiquity. R-M269 has been found, for instance, at a rate of ~44% among remains dating from the 11th to 13th centuries at Punta Azul, in the Canary Islands. These remains have been linked to the Bimbache (or Bimape), a subgroup of the Guanche.[62] In living males, it peaks in parts of North Africa, especially Algeria, at a rate of 10%.[63] In Sub-Saharan Africa, R-M269 appears to peak in Namibia, at a rate of 8% among Herero males.[64] In western Asia, R-M269 has been reported in 40% of Armenian males and over 35% in Turkmen males.[65][66] (The table below lists in more detail the frequencies of M269 in regions in Asia, Europe, and Africa.)
Apart from basal R-M269* which has not diverged, there are (as of 2017) two primary branches of R-M269:
R-L23 (Z2105/Z2103; a.k.a. R1b1a1b1) has been reported among the peoples of the Idel-Ural (by Trofimova et al. 2015): 21 out of 58 (36.2%) of Burzyansky District Bashkirs, 11 out of 52 (21.2%) of Udmurts, 4 out of 50 (8%) of Komi, 4 out of 59 (6.8%) of Mordvins, 2 out of 53 (3.8%) of Besermyan and 1 out of 43 (2.3%) of Chuvash were R1b-L23.[67]
Subclades within the paragroup R-M269(xL23) – that is, R-M269* and/or R-PF7558 – appear to be found at their highest frequency in the central Balkans, especially Kosovo with 7.9%, North Macedonia 5.1% and Serbia 4.4%.[41] Unlike most other areas with significant percentages of R-L23, Kosovo, Poland and the Bashkirs of south-east Bashkortostan are notable in having a high percentage of R-L23 (xM412) – at rates of 11.4% (Kosovo), 2.4% (Poland) and 2.4% south-east Bashkortostan.[41] (This Bashkir population is also notable for its high level of R-M73 (R1b1a1a1), at 23.4%.[41]) Five individuals out of 110 tested in the Ararat Valley of Armenia belonged to R-M269(xL23) and 36 to R-L23*, with none belonging to known subclades of L23.[68]
In 2009, DNA extracted from the femur bones of 6 skeletons in an early-medieval burial place in Ergolding (Bavaria, Germany) dated to around AD 670 yielded the following results: 4 were found to be haplogroup R1b with the closest matches in modern populations of Germany, Ireland and the USA while 2 were in Haplogroup G2a.[69]
The following gives a summary of most of the studies which specifically tested for M269, showing its distribution (as a percentage of total population) in Europe, North Africa, the Middle East and Central Asia as far as China and Nepal.
The phylogeny of R-M269 according to ISOGG 2017:
| M269/PF6517 |
| |||||||||||||||||||||||||||||||||||||||||||||
R1b1b (PF6279/V88; previously R1b1a2) is defined by the presence of SNP marker V88, the discovery of which was announced in 2010 by Cruciani et al.[44] Apart from individuals in southern Europe and Western Asia, the majority of R-V88 was found in the Sahel, especially among populations speaking Afroasiatic languages of the Chadic branch.
Based on a detailed phylogenic analysis, D'Atanasio et al. (2018) proposed that R1b-V88 originated in Europe about 12,000 years ago and crossed to North Africa between 8000 and 7000 years ago, during the 'Green Sahara' period. R1b-V1589, the main subclade within R1b-V88, underwent a further expansion around 5500 years ago, likely in the Lake Chad Basin region, from which some lines recrossed the Sahara to North Africa.[70]
Marcus et al. (2020) provide strong evidence for this proposed model of North to South trans-Saharan movement: The earliest basal R1b-V88 haplogroups are found in several Eastern European Hunter Gatherers close to 11,000 years ago. The haplogroup then seemingly spread with the expansion of Neolithic farmers, who established agriculture in the Western Mediterranean by around 7500 BP. R1b-V88 haplogroups were identified in ancient Neolithic individuals in Germany, central Italy, Iberia, and, at a particularly high frequency, in Sardinia. A part of the branch leading to present-day African haplogroups (V2197) was already derived in Neolithic European individuals from Spain and Sardinia, providing further support for a North to South trans-Saharan movement.[71][72][73] European autosomal ancestry, mtDNA haplogroups, and lactase persistence alleles have also been identified in African populations that carry R1b-V88 at a high frequency, such as the Fulani and Toubou.[74][75][72][76] The presence of European Neolithic farmers in Africa is further attested by samples from Morocco dating from c. 5400 BC onwards.[77][78]
Studies in 2005–08 reported "R1b*" at high levels in Jordan, Egypt and Sudan.[79][64][80][note 1] Subsequent research by Myres et al. (2011) indicates that the samples concerned most likely belong to the subclade R-V88. According to Myres et al. (2011), this may be explained by a back-migration from Asia into Africa by R1b-carrying people.[41][note 2]
Contrary to other studies, Shriner & Rotimi (2018) associated the introduction of R1b into Chad with the more recent movements of Baggara Arabs.[81]
| Region | Population | Country | Language | Sample size |
Total % |
R1b1b (R-V88) |
R1b1a1b (R-M269) |
R1b1b* (R-V88*) |
R1b1b2a2a1 (R-V69) |
|---|---|---|---|---|---|---|---|---|---|
| N Africa | Composite | Morocco | AA | 338 | 0.0% | 0.3% | 0.6% | 0.3% | 0.0% |
| N Africa | Mozabite Berbers | Algeria | AA/Berber | 67 | 3.0% | 3.0% | 0.0% | 3.0% | 0.0% |
| N Africa | Northern Egyptians | Egypt | AA/Semitic | 49 | 6.1% | 4.1% | 2.0% | 4.1% | 0.0% |
| N Africa | Berbers from Siwa | Egypt | AA/Berber | 93 | 28.0% | 26.9% | 1.1% | 23.7% | 3.2% |
| N Africa | Baharia | Egypt | AA/Semitic | 41 | 7.3% | 4.9% | 2.4% | 0.0% | 4.9% |
| N Africa | Gurna Oasis | Egypt | AA/Semitic | 34 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| N Africa | Southern Egyptians | Egypt | AA/Semitic | 69 | 5.8% | 5.8% | 0.0% | 2.9% | 2.9% |
| C Africa | Songhai | Niger | NS/Songhai | 10 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| C Africa | Fulbe | Niger | NC/Atlantic | 7 | 14.3% | 14.3% | 0.0% | 14.3% | 0.0% |
| C Africa | Tuareg | Niger | AA/Berber | 22 | 4.5% | 4.5% | 0.0% | 4.5% | 0.0% |
| C Africa | Ngambai | Chad | NS/Sudanic | 11 | 9.1% | 9.1% | 0.0% | 9.1% | 0.0% |
| C Africa | Hausa | Nigeria (North) | AA/Chadic | 10 | 20.0% | 20.0% | 0.0% | 20.0% | 0.0% |
| C Africa | Fulbe | Nigeria (North) | NC/Atlantic | 32 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| C Africa | Yoruba | Nigeria (South) | NC/Defoid | 21 | 4.8% | 4.8% | 0.0% | 4.8% | 0.0% |
| C Africa | Ouldeme | Cameroon (Nth) | AA/Chadic | 22 | 95.5% | 95.5% | 0.0% | 95.5% | 0.0% |
| C Africa | Mada | Cameroon (Nth) | AA/Chadic | 17 | 82.4% | 82.4% | 0.0% | 76.5% | 5.9% |
| C Africa | Mafa | Cameroon (Nth) | AA/Chadic | 8 | 87.5% | 87.5% | 0.0% | 25.0% | 62.5% |
| C Africa | Guiziga | Cameroon (Nth) | AA/Chadic | 9 | 77.8% | 77.8% | 0.0% | 22.2% | 55.6% |
| C Africa | Daba | Cameroon (Nth) | AA/Chadic | 19 | 42.1% | 42.1% | 0.0% | 36.8% | 5.3% |
| C Africa | Guidar | Cameroon (Nth) | AA/Chadic | 9 | 66.7% | 66.7% | 0.0% | 22.2% | 44.4% |
| C Africa | Massa | Cameroon (Nth) | AA/Chadic | 7 | 28.6% | 28.6% | 0.0% | 14.3% | 14.3% |
| C Africa | Other Chadic | Cameroon (Nth) | AA/Chadic | 4 | 75.0% | 75.0% | 0.0% | 25.0% | 50.0% |
| C Africa | Shuwa Arabs | Cameroon (Nth) | AA/Semitic | 5 | 40.0% | 40.0% | 0.0% | 40.0% | 0.0% |
| C Africa | Kanuri | Cameroon (Nth) | NS/Saharan | 7 | 14.3% | 14.3% | 0.0% | 14.3% | 0.0% |
| C Africa | Fulbe | Cameroon (Nth) | NC/Atlantic | 18 | 11.1% | 11.1% | 0.0% | 5.6% | 5.6% |
| C Africa | Moundang | Cameroon (Nth) | NC/Adamawa | 21 | 66.7% | 66.7% | 0.0% | 14.3% | 52.4% |
| C Africa | Fali | Cameroon (Nth) | NC/Adamawa | 48 | 20.8% | 20.8% | 0.0% | 10.4% | 10.4% |
| C Africa | Tali | Cameroon (Nth) | NC/Adamawa | 22 | 9.1% | 9.1% | 0.0% | 4.5% | 4.5% |
| C Africa | Mboum | Cameroon (Nth) | NC/Adamawa | 9 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| C Africa | Composite | Cameroon (Sth) | NC/Bantu | 90 | 0.0% | 1.1% | 0.0% | 1.1% | 0.0% |
| C Africa | Biaka Pygmies | CAR | NC/Bantu | 33 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| W Africa | Composite | — | 123 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| E Africa | Composite | — | 442 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| S Africa | Composite | — | 105 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| Total | 1822 | ||||||||
| V88 |
| ||||||||||||
Two branches of R-V88, R-M18 and R-V35, are found almost exclusively on the island of Sardinia.
As can be seen in the above data table, R-V88 is found in northern Cameroon in west central Africa at a very high frequency, where it is considered to be caused by a pre-Islamic movement of people from Eurasia.[64][82]
R1b1b1 is a sub-clade of R-V88, which is defined by the presence of SNP marker M18.[7] It has been found only at low frequencies in samples from Sardinia[60][83] and Lebanon.[84]
R1b2 is extremely rare and defined by the presence of PH155.[36] Living males carrying subclades of R-PH155 have been found in Bahrain, Bhutan, Ladakh, Tajikistan, Turkey, Xinjiang, and Yunnan. ISOGG (2022) cites two primary branches: R-M335 (R1b2a) and R-PH200 (R1b2b).
The defining SNP of R1b2a, M335, was first documented in 2004, when an example was discovered in Turkey, though it was classified at that time as R1b4.[34] Other examples of R-M335 have been reported in a sample of Hui from Yunnan, China[85] and in a sample of people from Ladakh, India.[86]
Spytihněv I, Duke of Bohemia, DNA testing on his remains suggests that his Y-haplogroup was R1b.[89]
The House of Bourbon, which has ruled as kings in France, Spain, and other European countries, have the R1b1b haplogroup.[90]
DNA testing on several mummies from the 18th dynasty of Egypt found haplogroup R1b. The mummy of Tutankhamun had the Y-haplogroup R1b and the mtDNA haplogroup K. He inherited this Y-haplogroup from his father, the KV55 mummy believed by many to be Akhenaten, and his grandfather, Amenhotep III, whose mummy was found entombed at KV35 with numerous relatives.[91][92]
Studies have shown that haplogroup R1b could have a protective effect on the immune system.[93] However, later studies have confirmed that the Y-chromosome has a very limited effect on coronary artery disease (CAD), for example, and that the previously purported link between Y-chromosome haplogroups and health is far from established scientifically.[94]
Seamless Wikipedia browsing. On steroids.
Every time you click a link to Wikipedia, Wiktionary or Wikiquote in your browser's search results, it will show the modern Wikiwand interface.
Wikiwand extension is a five stars, simple, with minimum permission required to keep your browsing private, safe and transparent.