Haplogroup N-M231
Human Y chromosome DNA grouping common in North Eurasia From Wikipedia, the free encyclopedia
Haplogroup N (M231) is a Y-chromosome DNA haplogroup defined by the presence of the single-nucleotide polymorphism (SNP) marker M231.[Phylogenetics 1]
This article possibly contains original research. (November 2021) |
| Haplogroup N | |
|---|---|
 | |
| Possible time of origin | 36,800 [95% CI 34,300–39,300] years before present (YFull[1]) 44,700 or 38,300 ybp depending on mutation rate[2] 41,900 [95% CI 40,175-43,591] ybp[3] |
| Coalescence age | 21,700 [95% CI 19,500–23,900] ybp (YFull[1]) 25,313 [95% CI 21,722–28,956] ybp[3] |
| Possible place of origin | Northern East Asia[4][5] |
| Ancestor | NO |
| Defining mutations | M231 |
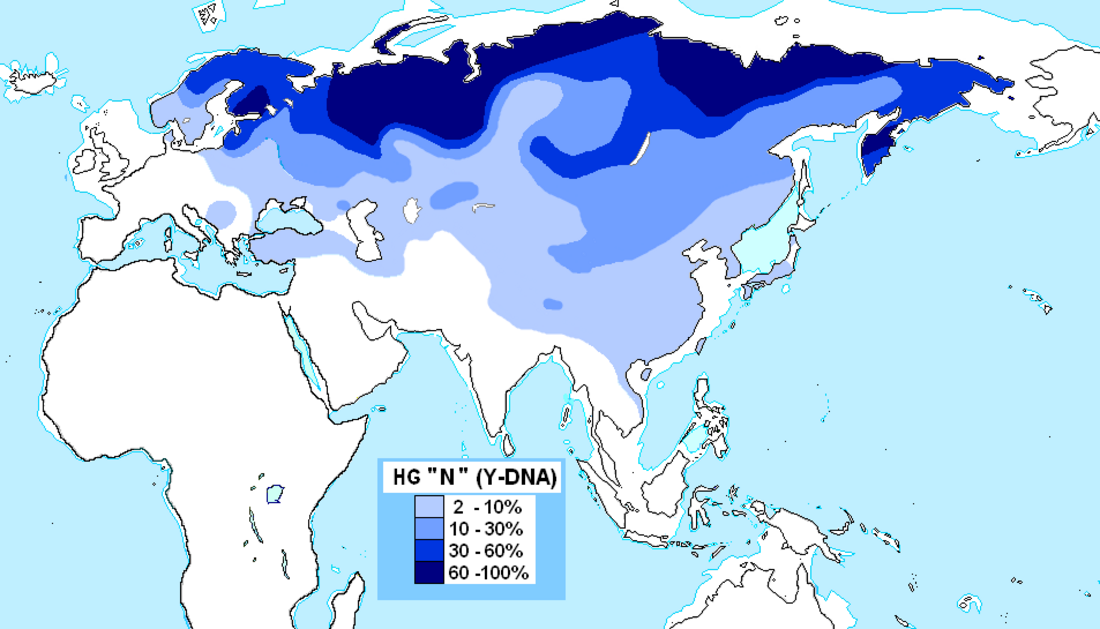
| Highest frequencies | Nganassan 58%[3]–94.1%,[6] Yakuts 81.8%[7]–94.6%,[8] Khakass (Shirinsky district) 90.2%,[9] Siberian Tatars 33.5% (Zabolotnie 89.5%, Iskero-Tobol 23.5%, Tatar-Bukharans 16.5%, Yalutorov 16.3%, Ishtyak-Tokuz 14.5%[10]), Ugrians 77.8%[3] (Khanty 64.3%[11]–89.3%,[12] Mansi 76%[12]), Udmurts 77.8%,[3] Khakas 41%[3]–65%,[11] Komi 33.3%[11]–79.5%,[6] Nenets 75%–92.9%[3] (Tundra Nenets 97.9%,[6] Forest Nenets 98.8%[6]), Vepsians 55%,[3] Finns 42.6% (West)[13]–70.9% (East)[13] or approx. 54%[3]–58.8%,[14] Tuvans 27.2–54.5%,[citation needed] Nanai 46.2%[3][15][16] (20% Hezhe in the PRC,[15] 44.6% Nanai in Russia,[3] 83.8% members of the Samar clan in the Gorin area of the Khabarovsk Territory[16]), Karelians 37.1%[5]–53.8%,[13] Arkhangelsk Russians 42.6% (Arkhangelsk 44.3%,[3] Pinega 40.8%[3]), Lithuanians 40.5%[3]–44.5%,[13] Latvians approx. 42% (41.6%,[13] 42.1%,[17] 43.0%[3]), Mari 41.2%,[3] Saami 40%, Chuvash 33.7%[14]–36%,[3] Buryats 34.5% (20.2%,[18] 25.0%,[19] 30.9%,[20] 48.0%[21]>), Koryaks 33.3%,[6] Estonians 30.6%[3]-33.9%,[13] Volga Tatars 27.8%,[3] Teleuts 25.0%,[6] Northern Altaians 21.8% (18.0%[11][22]–24.6%[23]), Pskov Russians 22.7%,[3] Russians 20%[24] Bashkirs 17.3%,[3] Sibe 17.1%[15]–18.0%,[25] Mordvins 12.5% (10%[3]–13.3%[3]), Mongols 11%,[26][20][15][19][27][28] Kalmyks 10.4% (Torguud 3.4%, Derbet 5.1%, Buzava 5.3%, Khoshut 38.2%),[29][28] Manchus 10% (5.8%,[20] 8.1%,[30] 9.1%,[25] 11.6%,[25] 12.5%,[25] 14.3%[15]), Belarusians 9.7%,[3] Central-Southern Russians 9.1% (Tver 13.2%,[31] Kursk 12.5%[31]–13.3%,[3] Belgorod 11.9%,[3] Kostroma 11.8%,[3] Smolensk 7.0%,[3] Voronezh 6.3%,[3] Oryol 5.5%[3]), Ukrainians 9.0%,[3] Southern Altaians 7.1% (4.2%[23]-9.7%[11]), Mulam 7.1%,[32] Sweden 6.8%[14] (0% Västra Götaland, Halland, Malmö, and Jönköping[33] - 19.5% Västerbotten[34]), Han Chinese 6.77% (0% to 21.4%),[25] Koreans 6.58% (4.41% to 12%) 12% Koreans,[35] 6.58% Koreans from KPGP(Korean Genome Project),[36] 6.9% Jeju[19] 6.4% Gochang [37] 6.3% Gangwon [19] 5.7% North Korean [38] 4.8% Gyeongsang,[19] 4.4% Jeolla,[19] 4.2% Chungcheong,[19] 4.0% Seoul,[39] 3.0% Daejeon,[39] 1.8% Seoul-Gyeonggi,[19] Ulchi 5.8%,[40] Tibetans 5.65%,[41] Kazakhs 5.33% [42] (Suan 0%, Qangly 0%, Oshaqty 0%, Jetyru 1.2%, Dulat 1.6%, Argyn 2.0%, Alimuly 2.5%, Ysty 3.5%, Baiuly 3.9%, Alban 4.3%, Qongyrat 7.4%, Qypshaq 10.3%, Jalair 10.9%, Qozha 16.7%, Syrgeli 65.6%), Northern Thai 5.2%,[43] Uyghurs 4.89% (2.8%,[44] 4.8%,[25] 4.99%,[45] 6.0%,[20] 8.6%[15]), Kyrgyz 3.9% (2.8% Kyzylsu,[46] 3.3% Kyzylsu,[47] 4.5% Kyrgyzstan,[27] 10% Urumqi[46]), Vietnamese 3.4%, Japanese 1.9% (0%,[2] 0.8%,[48] 0.9%,[49] 1.7%,[50] 2.5%,[19] 4.3%,[51] 4.8%,[20] 6.4%[15]) |
It is most commonly found in males originating from northern Eurasia. It also has been observed at lower frequencies in populations native to other regions, including parts of Central Asia, East Asia, and Southeast Asia.
However, the basal paragroup N* has only been found in populations indigenous to China and Cambodia.[4] Subclades of N-M231 have been found at low levels in Southeast Asia, the Pacific Islands, Southwest Asia and the Balkans. These factors tend to suggest that it originated in East Asia or Southeast Asia.
Origins

Haplogroup NO-M214 – its most recent common ancestor with its sibling, haplogroup O-M175 – is estimated to have existed about 36,800–44,700 years ago.[1][53][2]
It is generally considered that N-M231 arose in East Asia approximately 19,400 (±4,800) years ago and populated northern Eurasia after the Last Glacial Maximum. Males carrying the marker apparently moved northwards as the climate warmed in the Holocene, migrating in a counter-clockwise path, to eventually become concentrated in areas as far away as Fennoscandia and the Baltic.[5] The apparent dearth of haplogroup N-M231 amongst Native American peoples indicates that it spread after Beringia was submerged,[54] about 11,000 years ago.
Distribution
Summarize
Perspective

Haplogroup N has a wide geographic distribution throughout northern Eurasia, and it also has been observed occasionally in other areas, including Central Asia and the Balkans.
It has been found with greatest frequency among indigenous peoples of Russia, including Uralic peoples (Mari, Udmurt, Komi, Khanty, Mansi, Nenets, Nganasans), Turkic peoples (Yakuts, Dolgans, Khakasses, Tuvans, Tatars, Chuvashes, etc.), Buryats, Tungusic peoples (Evenks, Evens, Negidals, Nanais, etc.), Yukaghirs, Luoravetlans (Chukchis, Koryaks), and Siberian Eskimos, but certain subclades are very common in Finland, Estonia, Latvia, and Lithuania, and other subclades are found at low frequency in China (Yi, Naxi, Lhoba, Han Chinese, etc.).[55] Especially in ethnic Finnic peoples and Baltic-speaking peoples of northern Europe, the Ob-Ugric-speaking and Northern Samoyed peoples of western Siberia, and Turkic-speaking peoples of Russia (especially Yakuts,[citation needed],but also Altaians, Shors, Khakas, Chuvashes, Tatars, and Bashkirs). Nearly all members of haplogroup N among these populations of northern Eurasia belong to subclades of either haplogroup N-CTS6128/M2048 or haplogroup N-P43.
Y-chromosomes belonging to N1b-F2930/M1881/V3743, or N1*-CTS11499/L735/M2291(xN1a-F1206/M2013/S11466), have been found in China (where they account for approximately 3.62% of all Y-DNA[56]) and sporadically throughout other parts of Eurasia. The N-CTS6128/M2048 and N-P43 subclades of N1a-F1206/M2013/S11466 are found in high numbers in Northern Eurasia; however, members of N1a-F1206(xCTS6128, P43) are currently found mainly in northern China and Korea.
N2-Y6503, the other primary subclade of haplogroup N, is extremely rare and is mainly represented among extant humans by a recently formed subclade that is virtually restricted to the countries making up the former Yugoslavia (Bosnia-Herzegovina, Croatia, Serbia, and Montenegro), Hungary and Austria. Other members of N2-Y6503 include a Hungarian with recent ancestry from Suceava in Bukovina, a Slovakian, a few British individuals, and an Altaian.[1]
N* (M231)
Y-chromosomes that display the M231 mutation that defines Haplogroup N-M231, but do not display the CTS11499, L735, M2291 mutations that define Haplogroup N1 are said to belong to paragroup N-M231*.[4]
N-M231* has been found at low levels in China.[4] Out of a sample of 165 Han males from China, two individuals (1.2%) were found to belong to N*.[57] One originated from Guangzhou and one from Xi'an.[citation needed]
Among the ancient samples from the Baikal Early Neolithic Kitoi culture, one of the Shamanka II samples (DA250), dated to c. 6500 BP, was analyzed as NO1-M214 in the original study.[58] However, this same specimen (DA250 or Shamanka 250) has subsequently been found to belong to N-FT210118, the same clade as the other haplogroup N specimens from the same site (besides DA247, who belongs to N-Y147969). N-FT210118 is derived from N-L666/N-F2199 but basal to N-CTS6380, this latter being the most recent common ancestor of present-day N-P43 (found mainly among Maris, Udmurts, Komis, Chuvashes, Tatars, Nenets, Nganasans, Khanty, Mansi, Khakas, Tuvans, etc.) and N-F1101 (found mainly among East Asians). Furthermore, N-FT210118 has not been found in any living individual who has had his Y-DNA tested to date, and the estimated TMRCA of N-CTS6380 exceeds the estimated date of deposition of any of the specimens from the Shamanka site associated with the Kitoi culture, so it appears that the representatives of the Kitoi culture at Shamanka (or at least their Y-DNA) have gone extinct rather than being direct ancestors of any living people.[59][60]
N1 (CTS11499, Z4762, CTS3750)
In 2014, there was a major change in the definition of subclade N1, when LLY22g was retired as the main defining SNP for N1 because of reports of LLY22g's unreliability. According to ISOGG, LLY22g is problematic because it is a "palindromic marker and can easily be misinterpreted."[4] Since then, the name N1 has been applied to a clade marked by a great number of SNPs, including CTS11499, Z4762, and CTS3750. N1 is the most recent common ancestor of all extant members of Haplogroup N-M231 except members of the rare N2-Y6503 (N2-B482) subclade. The TMRCA of N1 is estimated to be 18,000 years before present (16,300–19,700 BP; 95% CI).[1] Since the revision of 2014, the position of many examples of "N1-LLY22g" within haplogroup N have become unclear. Therefore, it is better to check yfull and ISOGG 2019 in order to understand the updated structure of N-M231.
However, in older studies, N-LLY22g has been reported to reach a frequency of up to 30% (13/43) among the Yi people of Butuo County, Sichuan in Southwest China.[20][61][62] It is also found in 34.6% of Lhoba people.[62] N1-LLY22g* has been found in samples of Han Chinese, but with widely varying frequency:
- 6.8% (3/44) Han from Xi'an[20][61]
- 6.7% (2/30) Han from Lanzhou[15]
- 3.6% (3/84) Taiwanese Han[20]
- 2.9% (1/34) Han from Chengdu[15]
- 2.9% (1/35) Han from Harbin[15]
- 2.9% (1/35) Han from Meixian district[15]
- 0% (0/32) Han from Yining City[15]
Other populations in which representatives of N1*-LLY22g have been found include:
- Hani people (4/34 = 11.8%)[15]
- Sibe people (4/41 = 9.8%)[15]
- Tujia people (2/49 = 4.1%)[20]
- Manchu people (2/52 = 3.8%[20] to 2/35 = 5.7%[15]
- Bit people (1/28 = 3.6%)[63]
- Uyghurs (2/70 = 2.9%[15] to 2/67 = 3.0%)[20]
- Tibetan people (3/105 = 2.9%[20] to 3/35 = 8.6%)[15]
- Koreans (0/106 = 0.0% – 2/25 = 8%[5][15][64]
- Vietnamese people (2/70 = 2.9%)[20]
- Japanese people (0/70 Tokushima – 2/26 = 7.7% Aomori)[20]
- Manchurian Evenks (0/26 = 0.0%[15] to 1/41 = 2.4%)[20]
- Altai people (0/50 Northern to 5/96 = 5.2% Southern, or 0/43 Beshpeltir to 5/46 = 10.9% Kulada),[20][22]
- Shors (2/23 = 8.7%)[5]
- Khakas people (5/181 = 2.8%)[5]
- Tuvans (5/311 = 1.6%)[5]
- Southern Borneo (1/40 = 2.5%)[5]
- Forest Nenets (1/89 = 1.1%)[5]
- Yakuts (0/215 – 1/121 = 0.8%)[5]
- Turkish people (1/523 = 0.2%)[5] In Turkey, the total of subclades of haplogroup N-M231 amounts to 4% of the male population.
- One individual who belongs either to N* or N1* has been found in a sample of 77 males from Kathmandu, Nepal (1/77 = 1.3% N-M231(xM128,P43,Tat))[65]
N1(xN1a,N1c) was found in ancient bones of Liao civilization:[66]
- Niuheliang (Hongshan Culture, 6500–5000 BP) 66.7%(=4/6)
- Halahaigou (Xiaoheyan Culture, 5000–4200 BP) 100.0%(=12/12)
- Dadianzi (Lower Xiajiadian culture, 4200–3600 BP) 60.0%(=3/5).
N-CTS4309: two people identified with this subgroup in Iraq. Very rare.
N1a (F1206/M2013/S11466)
The N1a2-F1008/L666 clade and N1a1-M46/Page70/Tat are estimated to share a most recent common ancestor in N1a-F1206/M2013/S11466 approximately 15,900 [95% CI 13,900 <-> 17,900] years before present[1] or 17,621 [95% CI 14,952 <-> 20,282] years before present.[3]
N1a1 (M46/Page70/Tat, L395/M2080)
All M46 in Yfull database are M178, being a quarter younger than separation from F1139.[67]
The mutations that define the subclade N-M46[Phylogenetics 2] are M46/Tat and P105. This is the most frequent subclade of N. It probably arose in a Northeast Asian population, because the oldest ancient samples comply with this genetic profile.[68] N has experienced serial bottlenecks in Siberia and secondary expansions in eastern Europe.[5] Haplogroup N-M46 is approximately 14,000 years old.
In Siberia, haplogroup N-M46 reaches a maximum frequency of approximately 90% among the Yakuts, a Turkic people who live mainly in the Sakha (Yakutia) Republic. However, N-M46 is present with much lower frequency among many of the Yakuts' neighbors, such as Evenks and Evens.[8] It also has been detected in 5.9% (3/51) of a sample of Hmong Daw from Laos,[63] 2.4% (2/85) of a sample from Seoul, South Korea,[26] and in 1.4% (1/70) of a sample from Tokushima, Japan.[20] Y-DNA haplogroup N-M46 accounts for about 1.07% of all Y-DNA in present-day China;[69] most of those (about 0.74% of all present-day Chinese males[70]) belong to the basal N-F4063 subclade, but the N-M2058 > N-A9408 > N-Y77895 > N-Y70200 subclade (accounting for about 0.15% of all present-day Chinese males[71]) and the N-CTS11808 > N-CTS10336 > N-Y6058 > N-Y16323 > N-F4205 subclade (accounting for about 0.07% of all present-day Chinese males[72]) are also readily detectable. Notably, China also has a significant presence (accounting for approximately 0.51% of all present-day Chinese males[73]) of N-MF14176, which is derived from N-F2584 but basal to N-M46.
The haplogroup N-M46 has a low diversity among Yakuts suggestive of a population bottleneck or founder effect.[74] This was confirmed by a study of ancient DNA which traced the origins of the male Yakut lineages to a small group of horse-riders from the Cis-Baikal area.[75]
N-Tat has been observed with greatly varying frequency in samples from Sweden. Karlsson et al. (2006) found N-Tat in 44.7% (17/38) of a sample of Saami nomads from Jokkmokk, 19.5% (8/41) of a sample from Västerbotten, 14.5% (8/55) of a sample from Uppsala, 10.0% (4/40) of a sample from Gotland, 9.5% (4/42) of a sample from Värmland, 7.3% (3/41) of a sample from Östergötland/Jönköping, 2.4% (1/41) of a sample from Blekinge/Kristianstad, and 2.2% (1/45) of a sample from Skaraborg.[34]
Lappalainen et al. (2008) found N-Tat in 14.4% (23/160) of a sample from Sweden.[13]
Lappalainen et al. (2009) found N-Tat in 15.4% (4/26) of a sample from Södermanland, 12.5% (3/24) of a sample from Västmanland, 12.1% (4/33) of a sample from Uppsala, 7.8% (4/51) of a sample from Gothenburg, 7.0% (3/43) of a sample from Norrbotten, 6.8% (5/73) of a sample from Skåne, 6.6% (15/228) of a sample from Stockholm, 6.3% (3/48) of a sample from Sydnorrland, 6.3% (2/32) of a sample from Västerbotten, 6.3% (2/32) of a sample from Örebro, 5.9% (3/51) of a sample from Värmland/Dalarna, 5.4% (2/37) of a sample from Östra Götaland, and 5.1% (2/39) of a sample from southeastern Sweden (Kalmar, Gotland, Kronoberg, and Blekinge). They did not find any instance of N-Tat in their samples from Jönköping (0/28), Malmö (0/29), Halland (0/34), or Västra Götaland (0/75).[33]
N1a1a (M178)
The subclade N-M178[Phylogenetics 3] is defined by the presence of markers M178 and P298. N-M178* has higher average frequency in Northern Europe than in Siberia, reaching frequencies of approximately 60% among Finns and approximately 40% among Latvians, Lithuanians & 35% among Estonians.[76][13]
Miroslava Derenko and her colleagues noted that there are two subclusters within this haplogroup, both present in Siberia and Northern Europe, with different histories. The one that they labelled N3a1 first expanded in south Siberia and spread into Northern Europe. Meanwhile, the younger subcluster, which they labelled N3a2, originated in south Siberia (probably in the Baikal region).[76]
N-M178 was also found in two Na-Dené speaking Tłı̨chǫs in North America.[77]
Neolithic samples from Baikal area have yielded plenty of yDNA N specimens, and one sample from Fofonovo, Buryatia, 5000-4000 BC is among the first Tat samples in the ancient record.[68]
Earliest samples of N1a1a-L708 were found in Trans-Baikal (brn008, N1a1a1*-L708; brn003, N1a1a1a1*-M2126) between 8,000 and 6,000 YBP. Downstream samples were found in Yakutia (N4b2, N1a1a1a1a*-Z1979) and Krasnoyarsk Krai (kra001, N1a1a1a1a*-L392), between 5,000 and 4,000 YBP.[78][79]
N1a2 (F1008/L666)
N1a2a-M128 and N1a2b-B523/P43 are estimated to share a most recent common ancestor in N1a2-F1008/L666 approximately 8,700 [95% CI 7,500 <-> 9,900] years before present,[1] 9,314 [95% CI 7,419 <-> 11,264] years before present,[3] or 10,430 years before present.[80]
At least three of six tested male specimens from the Early Neolithic (ceramic-using hunter-gatherer of approximately 7200–6200 years ago) layer at the Shamanka archaeological site near the southern end of Lake Baikal have been found to belong to N1a2-L666.[58]
N1a2a-M128
| Haplogroup N-M128 | |
|---|---|
 | |
| Possible place of origin | Asia |
| Ancestor | N1a2 (F1008/L666) |
| Descendants | F710, F1998 |
| Defining mutations | M128 |
| Highest frequencies | Manchus about 1% (0/43 Bijie,[81] 0/30,[19] 30/2938 = 1.0% China total[82] 1/52 = 1.9% Liaoning,[20] 2/35 = 5.7% Xiuyan Manchu Autonomous County),[15] Koreans 0/43 South Korea,[15] 0/25 China,[15] 1/75 = 1.3% South Korea,[20] Xibe 1/41 = 2.4%,[15] Han 0.89% (Shanxi 1.83%, Henan 1.53%, Gansu 1.46%, Hebei 1.41%, Shaanxi 1.40%, Tianjin 1.38%, Shandong 1.17%, Beijing 1.14%, Inner Mongolia 1.13%, Heilungjiang 1.12%, Liaoning 1.12%, Fujian 1.12%, Anhui 0.93%, Jilin 0.86%, Ningxia 0.82%, Jiangsu 0.81%, Yunnan 0.79%, Hubei 0.77%, Zhejiang 0.73%, Taiwan 0.70%, Sichuan 0.54%, Hunan 0.52%, Chongqing 0.51%, Guizhou 0.51%, Guangdong 0.45%, Hainan 0.43%, Xinjiang 0.43%, Shanghai 0.36%, Jiangxi 0.34%, Guangxi 0.23%, Qinghai 0.18%), Mongolians 0.7% (Liaoning 1.9%, Horqin 0.3%, Ulaanbaatar 0.0%), Japanese 0.3% (Tokushima 3.5%, Ibaraki 2.0%, Hiroshima 1.3%, Nagoya 1.0%, Osaka 0.2%, Tokyo 0.0%, Shizuoka 0.0%, Yamaguchi 0.0%), Kazakhs 0.1% (Khoja 1.9%, Baiuly 0.6%, Uysun 0.4%) |
This subclade is defined by the presence of the marker M128.[Phylogenetics 4] N-M128 was first identified in a sample from Japan (1/23 = 4.3%) and in a sample from Central Asia and Siberia (1/184 = 0.5%) in a preliminary survey of worldwide Y-DNA variation.[51] Subsequently, it has been found with low frequency in some samples of the Manchu people, Sibe people, Evenks, Koreans, Han Chinese, Hui, Tibetans, Vietnamese, Bouyei people, Kazakhs, Uzbeks, Uyghurs, Salars, Tu, Mongols, the Buzava tribe of Kalmyks,[29] Khakas, and Komis.[5] About 0.88% of all males in present-day China belong to N-M128 > N-F710, and it has been suggested that this clade may be associated with the aristocracy or even the royal family of the Zhou Dynasty.[83]
A number of a Han Chinese, an Ooled Mongol, a Qiang, and a Tibetan were found to belong to a sister branch (or branches) of N-M128 under paragroup N-F1154*.[84]
A neolithic sample brn002 (~5,940 BP) in Trans-Baikal was discovered to be an early offshoot upstream of N-M128.[85]
As a genetic testing result of Yelü clan, a royal family of the Liao Dynasty and Khitan descents, it was found to belong to N-F1998, a downstream of N-M128.
According to "The deep population history of northern East Asia from the Late Pleistocene to the Holocene",[86] the basal yDNA N-M128/mtDNA B5b2 HGDP01293 individual[87] occupied a position on the PCA between the Jiangsu Province’s and Anhui province’s specimens, but not far from the Shandong province’s mtDNA R11’B6>R11b specimen,[86] while a later descendant of the yDNA N-M128 clade, belonging to mtDNA R11’B6>R11b, was reported from the ancient DNA of the Western Zhou Cemetery, tomb M18.[88] Based on ancient DNA, the distribution of mtDNA B5b2 after 9500 years ago and prior to 4600 years ago in the direction of the Anhui and Jiangsu provinces from Shandong from the vicinity of the future Shandong Longshan Yinjiacheng site is shown in “Maternal genetic structure in ancient Shandong between 9500 and 1800 years ago”,[89] while the existence of the local Paleolithic Northern East Asian substratum, represented by individuals of the basal died-out yDNA O-M164*, separating from the Southern East Asian yDNA O-M188 and contributing to yDNA C2-M217 ancestors of Altaic and Korean representatives, was shown in "Human genetic history on the Tibetan Plateau in the past 5100 years".[90] Having occupied the position on the PCA between the Jiangsu Province’s and Anhui Province’s specimens, the basal yDNA N-M128/mtDNA B5b2 HGDP01293 individual became a participant of the uniform genetic cline, which spanned from Jiangsu and Anhui individuals to the Tai-speaking Dai people, and from Jiangsu and Anhui individuals to the ancient individual WGM20, belonging to mtDNA M11, of the Yangshao Wanggou site, dated to 5000-5500 years ago, and this ancient age also encompassed ancient yDNA pre-N-M128 Mazongshan individuals[91] and modern yDNA N-M128-affiliated Gansu Province’s individuals, who appeared to be included on the mentioned genetic cline closer to the ancient Henan province’s specimens of the Longshan period ca. 4000 years ago, than to the more genetically basal ancient individual WGM20 of the Yangshao Wanggou site, dated to 5000-5500 years ago.[86]
N1a2b (P43)
Haplogroup N-P43[Phylogenetics 5] is defined by the presence of the marker P43. Additionally, haplogroup N-P43 is defined by a marker Y3214, which is shared with a younger yDNA O1b2-K14, distributed in Japan (YFull). It has been estimated to be approximately 4,000 to 5,500 years old (TMRCA 4,510 years,[80] TMRCA 4,700 [95% CI 3,800 <-> 5,600] ybp,[1] or 4,727 [95% CI 3,824 <-> 5,693] years before present).[3] It has been found very frequently among Northern Samoyedic peoples, speakers of Ob-Ugric languages, and northern Khakassians, and it also has been observed with low to moderate frequency among speakers of some other Uralic languages, Turkic peoples, Mongolic peoples, Tungusic peoples, and Siberian Yupik people.
The highest frequencies of N-P43 are observed among north-west Siberian populations: 92% (35/38)[5] in a sample of Nganasan, 78% (7/9)[92][14] in a sample of Enets, 78% (21/27)[31] in a sample of Khants, 75% (44/59)[5] in a sample of Tundra Nenets, 69% (29/42)[3] in another sample of Nenets, 60% (15/25)[12] in a sample of Mansi, 57% (64/112)[11] in another sample of Khants, 54% (27/50)[3] in another sample of Nganasan, 45% (40/89)[5] in a sample of Forest Nenets, 38% (18/47)[93] in a third sample of Khants, and 25% (7/28)[12] in a fourth sample of Khants. In Europe, the N-P43 types have their highest frequency of 20% among Volga-Uralic populations. The extreme western border of the spread of N-P43 is Finland, where this haplogroup occurs only at marginal frequency – 0.4%. Yet N-P43 is quite frequent among Vepsas (17.9%), a small Finnic population living in immediate proximity to Finns, Karelians and Estonians.[5]
Haplogroup N-P43 also has been observed with very high frequency (26/29 = 89.7% of a sample from the settlement of Topanov and 19/22 = 86.4% of a sample from the settlement of Malyi Spirin) in samples of Kachins, a Turkic-speaking ethnic group or territorial subgroup of the Khakas people, from Shirinsky district of northern Khakassia.[9] There appears to be a cline through the Sagai (another Turkic-speaking ethnic group that is now considered to be a constituent of the Khakas people), with 46.2% (55/119) of Sagai sampled from Ust'–Es', Esino, Ust'–Chul', and Kyzlas settlements of Askizsky district of central Khakassia belonging to haplogroup N-P43 vs. only 13.6% (11/81) of Sagai sampled from Matur, Anchul', Bol'shaya Seya, and Butrakhty settlements of Tashtypsky district of southern Khakassia belonging to this haplogroup.[9] However, other researchers' samples of Khakas people have exhibited only moderate frequencies of N-P43 or potential N-P43. Derenko et al. (2006) examined a sample of Khakassians (n=53) collected in the settlements of Askiz, Shirinsk, Beisk and Ordzhonikidzevsk districts of Khakass Republic and found that 15 of them (28.3%) belonged to N-LLY22g(xTat).[18] Rootsi et al. (2007) examined a sample of Khakas (n=181) and found that 31 of them (17.1%) belonged to N-P43;[5] retested 174 of the individuals in this sample and found that 27 of them (15.5%) belonged to the N-B478 (Asian/northern Samoyedic) subclade of N-P43 and 2 of them (1.1%) belonged to the N-L1419 (European/Volga Finnic and Chuvash) subclade of N-P43 for a total of 29 (16.7%) N-P43.[3]
Haplogroup N-P43 forms two distinctive subclusters of STR haplotypes, Asian and European, the latter mostly distributed among Finno-Ugric-speaking peoples and related populations.[5]
N1a2b1-B478
The TMRCA of N-B478 has been estimated to be 3,007 [95% CI 2,171 <-> 3,970] years before present.[3] It is one of the most prevalent Y-DNA haplogroups among indigenous populations of northwestern Siberia: 69.0% (29/42) Nenets, 50.0% (25/50) Nganasan, 22.2% (12/54) Dolgan from Taymyr, 7.0% (3/43) Selkup, 1.6% (1/63) Ob-Ugrian. It is also quite prevalent among populations of Central Siberia, Southern Siberia, and Mongolia: 17.9% (17/95) Tuvan, 15.5% (27/174) Khakas, 13.0% (6/46) Tozhu Tuvans,[28] 8.7% (2/23) Shor, 8.3% (2/24) Even, 8.2% (5/61) Altaian, 5.3% (3/57) Evenk, 5.0% (19/381) Mongol, 4.9% (3/61) Sart-Kalmak (partial descendants of Oirat Mongols in Kyrgyzstan),[28] 4.2% (9/216) Yakut, 2.1% (1/47) Torgut (Mongolia),[28] 1.4% (1/69) Derbet (Kalmykia),[28] 0.9% (1/111) Buryat. A geographically outlying member has been found in a sample of Chuvash (1/114 = 0.88%).[3]
Karafet et al. (2018) have found N-P63, which appears to be roughly phylogenetically equivalent to N-B478, in 91.2% (31/34) Nganasan, 63.8% (30/47) Tundra Nenets, 42.7% (35/82) Forest Nenets, 14.0% (8/57) Dolgan, 7.0% (9/129) Selkup, 3.3% (3/91) Evenk, 2.7% (2/75) Mongol, 2.6% (2/78) Komi, 2.5% (2/80) Buryat, and 2.0% (2/98) Altai Kizhi.[6] This haplogroup was not observed in samples of Yukaghir (0/10), Koryak (0/11), Teleut (0/40), Ket (0/44), Yakut (0/62), or Khanty (0/165) populations.[6]
Kharkov et al. (2023) have found N-B478 in greatly differing percentages of samples of Khanty from two different villages of Khanty-Mansi Autonomous Okrug: 60.9% (39/64) of a sample of Khanty from the village of Russkinskaya, Surgut district and 14.8% (8/54) of a sample of Khanty from the village of Kazym, Beloyarsky district. Five of the eight members from the village of Kazym share a subclade marked by the B172 and Z35108 SNPs with all previously surveyed Nenets men from the Vanuito phratry belonging to the Vanuito, Puiko and Yaungat clans and the Purungui clan of Khanty origin.[94]
N1b (F2930)
Haplogroup N1b has been predominantly found in the Yi people, a Tibeto-Burman speaking ethnic group in southwestern China who originated from ancient Qiang tribes in northwestern China.[55] However, it also has been found in people all over China (where they account for approximately 3.62% of the country's male population and are mainly distributed in Shandong, Jiangsu, Zhejiang, Anhui, etc.[95]) and in some individuals from Spain,[59] Ecuador,[59] Poland,[59][1] Belarus,[59] Russia,[59][1] Iraq,[59] India,[59][1] Kazakhstan,[59] Korea,[59][1] Japan,[59][1] Bhutan,[59] Vietnam,[59][1] Cambodia,[59][1] Laos,[59] Thailand,[59][1] Malaysia,[1] and Singapore.[1]
N2 (Y6503)
N2 (Y6503/FGC28528; B482/FGC28394/Y6584) – a primary branch of haplogroup N-M231, is now represented mainly by a subclade, N-FGC28435, that has spread probably some time in the first half of the second millennium CE[96] and that has been found in individuals from Serbia, Croatia, Bosnia and Herzegovina, Montenegro, and Turkey (Istanbul).[97][96]
N-Y7310 (or N-F14667) subsumes N-FGC28435 and likewise probably descends from a common ancestor who has lived some time in the first half of the last millennium. However, members of N-Y7310(xFGC28435) exhibit a greater geographic range, including an individual from Rostov Oblast of Russia and a Romanian Hungarian individual with ancestry from Suceava, Bukovina.[1]
Other branches of N-P189 include members from Turkey,[1] Russia (Moscow Oblast),[1] France (Charente-Maritime),[1] and England (Devon).[1][59] The most recent common ancestor of all the aforementioned extant N-P189 lineages has been estimated to be 4,900 (95% CI 5,700 <-> 4,100) years before present.[1] An archaeological specimen attributed to the Botai culture of northern Kazakhstan and dated to the latter half of the fourth millennium BCE belongs to N-P189*, being basal to present-day European members of N-P189.[98][1]
Lineages that belong to N-Y6503(xP189) and are only distantly related (with a time to most recent common ancestor estimated to be greater than 10,000 years before present)[1] to the aforementioned members of N-P189 have been found in an individual from the present-day Altai Republic[1] and probably also in an archaeological specimen attributed to the Iron Age Mezőcsát culture of what is now Hungary (approx. 2,900 years before present)[99] and in an archaeological specimen attributed to the Kitoi culture of ceramic-using foragers of the area around Lake Baikal (approx. 6,700 years before present).[98]
Ancient peoples
A sample excavated at the Houtaomuga site in the Yonghe neighborhood of Honggangzi Township, Da'an, Jilin, China dating back to 7430–7320 years ago (Phase II of the Early Neolithic) has been found to belong to Y-DNA haplogroup N and mtDNA haplogroup B4c1a2. This sample is autosomally identical with the Neolithic Amur River Basin populations, of which Nivkh people are the closest modern representative. As the paper detected this ancestry in terminal Pleistocene USR1 specimen in Alaska, it is therefore, postulated that there was gene flow from Amur to America of a population belonging to a hypothetical Chukotko-Kamchatkan–Nivkh linguistic family.[citation needed]
N has also been found in many samples of Neolithic human remains exhumed from Liao civilization in northeastern China, and in the circum-Baikal area of southern Siberia.[66] It is suggested that yDNA N, reached southern Siberia from 12 to 14 kya. From there it reached southern Europe 8-10kya.[52]
Phylogeny
Summarize
Perspective
Phylogenetic tree
In the following tree the nomenclature of three sources is separated by slashes: ISOGG Tree 10 December 2017 (ver.12.317)
- NO-M214
- N-M231/Page91, M232/M2188
- N1-Z4762/CTS11499/L735/M2291
- N1a-L729
- N-Z1956
- N-Y149447 China (Shaanxi[1])
- N1a1-M46/Page70/Tat
- N1a1a-M178
- N1a1a1-F1419
- N1a1a1a-L708
- N1a1a1a1-P298/M2126 China (Urumqi, Kashgar Prefecture, Turpan, Aksu Prefecture, Xianyang, Jincheng, Kaifeng, Qiqihar[80])
- N1a1a1a1a-L392
- N1a1a1a1a1-CTS10760
- N1a1a1a1a1a-CTS2929/VL29 Found with high frequency among Lithuanians, Latvians, Estonians, northwestern Russians, Swedish Saami, Karelians, Nenetses, Finns, and Maris, moderate frequency among other Russians, Belarusians, Ukrainians, and Poles, and low frequency among Komis, Mordva, Tatars, Chuvashes, Dolgans, Vepsa, Selkups, Karanogays, and Bashkirs[3]
- N1a1a1a1a1a1-Z4908
- N-Y46443
- N-Y46443* Russia (Moscow Oblast[1])
- N-BY33095 Russia (Samara Oblast[1])
- N1a1a1a1a1a1a-L550/S431
- N-L550* Sweden (Kronoberg County[1])
- N-Y9454 Sweden (Västra Götaland County,[1] Örebro County[1]), Finland,[1] Russia[1]
- N-Y20911 Finland (Western Finland Province,[1] Southern Finland Province[1])
- N-Y7795
- N-Y7795* Sweden (Norrbotten County,[1] Södermanland County[1])
- N-Y29766 Norway (Hedmark[1])
- N-Y20918 Sweden (Östergötland County,[1] Västerbotten County[1]), Finland (Western Finland Province[1])
- N-Y28771
- N-Y61225 Sweden (Södermanland County,[1] Uppsala County[1])
- N-Y30126
- N-Y30126* Sweden (Östergötland County[1])
- N-Y29764
- N-Y29764* Sweden (Stockholm County[1])
- N-Y30123 Sweden (Västerbotten County[1]), Finland[1]
- N-S9378
- N-S9378* Sweden (Västra Götaland County[1])
- N-S18447 Sweden (Södermanland County,[1] Uppsala County[1])
- N-Y36282
- N-Y36282* Finland (Southern Finland Province[1]), Estonia (Ida-Virumaa[1])
- N-BY21957 Poland (Kujawsko-pomorskie[1]), Sweden (Stockholm County[1]), Russia (Pskov Oblast[1]), Finland (Southern Finland Province[1])
- N-FGC14542
- N-Y4341
- N-BY21874 Sweden (Södermanland County[1]), Finland[1]
- N-Y4338
- N-Y4338*
- N-Y4339 Sweden (medieval Sigtuna[1])
- N-Y12104
- N-Y19111
- N-Y57577 Sweden (Skåne County[1])
- N-Y22774 Finland (Western Finland Province[1])
- N-Y5611
- N-Y5611* England[1]
- N-Y21546 Sweden (Stockholm County,[1] Västmanland County[1])
- N-F1983 Sweden (Gävleborg County[1]), Russia (Lipetsk Oblast[1])
- N-Y10932
- N-Y85136 Sweden (Uppsala County[1])
- N-Y10931 Russia[1]
- N1a1a1a1a1a1a1-L1025/B215 Highest frequency among Lithuanians, significant in Latvians and Estonians and lesser frequency in Belarusians, Ukrainians, South-West Russians, and Poles. With exception of Estonians, L1025 has highest share among N-M231 clades in previously mentioned populations. Also observed in Finland and Sweden, with sporadic instances in Norway, Germany, Netherlands, United Kingdom, the Azores, Czech Republic, and Slovakia.
- N-L1025* Russia (Kursk Oblast[1])
- N-BY30389 Sweden (Västernorrland County[1]), Finland (Western Finland Province[1])
- N-Y13982
- N-Y13982* Portugal (Azores[1])
- N-Y31236 Lithuania (Marijampolė County,[1] Vilnius County[1])
- N-A11940
- N-Y140872 Sweden (Södermanland County,[1] Norrbotten County[1]), Finland (Southern Finland Province[1])
- N-Y143451 Russia (Bashkortostan[1])
- N-Y5580
- N-Y93996 Lithuania (Šiauliai County[1]), Poland (Podlaskie[1])
- N-BY158 Russia (Voronezh Oblast,[1] Smolensk Oblast,[1] Tatarstan[1]), Belarus (Vitebsk Region[1]), Lithuania (Vilnius County,[1] Šiauliai County,[1] Panevėžys County[1]), Poland (Podlaskie[1]), Ukraine (Poltava Oblast[1]), Kazakhstan (Jambyl Region[1])
- N-Z16975
- N-VL69 Belarus (Minsk Region[1]), Russia (Bryansk Oblast[1]), Kazakhstan (Kostanay Region[1])
- N-Z16976
- N-Z16976* Belarus (Grodno Region[1]), Ukraine (Khmelnytskyi Oblast[1])
- N-Y13475 Poland,[1] Lithuania[1]
- N-Y21578 Lithuania (Kaunas County,[1] Telšiai County[1]), Russia (Smolensk Oblast[1])
- N-Y6129 Poland (Warmińsko-mazurskie[1]), Lithuania[1]
- N-Y19113 Poland (Warmińsko-mazurskie[1]), Lithuania (Tauragė County[1])
- N-Y134492 Belarus (Grodno Region,[1] Minsk Region,[1] Mogilev Region[1]), Lithuania,[1] United States (Illinois[1])
- N-L551
- N-L551* Lithuania (Vilnius County[1])
- N-Y15251 Lithuania (Alytus County[1]), Poland (Podlaskie[1])
- N-Y46313 Latvia[1]
- N-Y86578 Russia (Belgorod Oblast[1])
- N-Y14152
- N-BY21911 Poland,[1] Latvia (Krāslava district[1]), Ukraine (Kyiv Oblast[1]), Finland (Southern Finland Province[1])
- N-Y13979 Lithuania,[1] Russia (Ryazan Oblast[1]), Germany[1]
- N-Y4706
- N-Y4706* Sweden (Stockholm County,[1] Södermanland County,[1] Västra Götaland County[1]), Germany (Lower Saxony[1]), Finland (Southern Finland Province[1]), Russia (Tatarstan[1])
- N-A705 Sweden (Kalmar County,[1] Östergötland County[1])
- N-BY21893 Poland (Wielkopolskie[1])
- N-Y139030 Sweden (Västerbotten County[1]), Norway (Østfold[1])
- N-Y183040 Russia (Vologda Oblast,[1] Tatarstan[1])
- N-Y4707 Finland (Western Finland Province,[1] Åland,[1] Southern Finland Province,[1] Eastern Finland Province,[1] Oulu Province[1])
- N-Z16981
- N-A2358=N-BY14135 England,[1] Finland,[1] Latvia (Daugavpils apriņķis[1])
- N-CTS8173 Estonia (Lääne-Virumaa[1])
- N-FT213922 [59])
- N-BY32524 Estonia (Järvamaa[1]), Finland (Oulu Province[1])
- N-FGC39882 Lithuania (Telšiai County[1])
- N-FT6082 [59])
- N-A11470 Russia (Tyumen Oblast[1]), Lithuania (Panevėžys County[1]), Finland (Southern Finland Province[1]), Netherlands (South Holland,[1] North Brabant[1])
- N-BY21926 Russia, Latvia, Belarus, Lithuania[59])
- N-Y15922=N-Z35238 Finland (Southern Finland Province[1]), Estonia (Ida-Virumaa,[1] Pärnumaa[1]), Latvia (Valmiera,[1] Liepāja,[1] Talsu apriņķis[1]), Russia (Pskov Oblast[1]), Poland (Pomorskie[1])
- N-Y6075 Poland (Małopolskie,[1] Mazowieckie[1]), Ukraine (Lviv Oblast[1]), Slovakia,[1] Czech Republic (Moravian-Silesian Region[1]), United States (New Jersey[1])
- N-Y11882
- N-Y11882* Russia (Tatarstan[1])
- N-ZS11617 Russia (Yaroslavl Oblast[1]), Lithuania[1]
- N-Y24601 Latvia[1]
- N-Y94659 Lithuania,[1] Belarus (Minsk Region[1])
- N-Y32725
- N-FT96305 Latvia (Krāslava district[1]), Estonia (Viljandimaa[1])
- N-Y33333 Ukraine (Sumy Oblast[1]), Belarus (Brest Region[1])
- N-FT213922 [59])
- N-Y46443
- N1a1a1a1a1a2-CTS9976
- N-L1022
- N-Y19098
- N-Y19098* Sweden (Västernorrland County[1])
- N-A12258 Finland (Eastern Finland Province[1])
- N-BY117178 Finland (Eastern Finland Province[1])
- N-Y5004
- N-Y7300
- N-A17632
- N-Y15813
- N-Y15813* Finland (Western Finland Province[1])
- N-Y49008 Finland (Western Finland Province,[1] Oulu Province[1])
- N-Y46886 Finland (Western Finland Province[1])
- N-Y15812
- N-A14187 Finland (Western Finland Province[1]), Estonia (Läänemaa[1])
- N-Y24617 Finland (Western Finland Province[1]), Sweden (Dalarna County[1]), Russia (Kursk Oblast[1])
- N-Y23576
- N-Y23576* Finland (Eastern Finland Province[1])
- N-Y15615
- N-Y5005
- N-Y22106
- N-Y47789 Finland (Western Finland Province[1])
- N-Y79341 Finland (Western Finland Province[1]), Sweden (Västra Götaland County,[1] Värmland County[1])
- N-Y10756
- N-A13656 Finland (Western Finland Province,[1] Lapland Province[1]), Sweden (Dalarna County[1])
- N-PH2196
- N-CTS11122 Finland (Eastern Finland Province[1])
- N-A17277 Finland (Western Finland Province,[1] Southern Finland Province[1])
- N-PH547 Finland (Lapland Province,[1] Eastern Finland Province,[1] Southern Finland Province[1]), Sweden (Västerbotten County,[1] Norrbotten County[1])
- N-Y5003
- N-Y5003* Estonia (Hiiumaa[1]), Finland (Southern Finland Province[1]), United Kingdom (Westminster[1])
- N-BY22001 Estonia (Läänemaa[1]), Finland (Western Finland Province[1]), Sweden (Norrbotten County[1])
- N-BY6007 Russia (Lipetsk Oblast[1])
- N-Y132182 Sweden (Värmland County[1]), Estonia (Viljandimaa[1])
- N-Y20917 Sweden (Kronoberg County[1]), Finland (Western Finland Province,[1] Southern Finland Province[1])
- N-Y24502 United Kingdom (Scottish Borders,[1] Calderdale,[1] Kirklees[1])
- N-Y18420 Finland (Southern Finland Province,[1] Western Finland Province[1])
- N-Z35267 Finland (Western Finland Province,[1] Åland Province,[1] Southern Finland Province[1]), Estonia (Pärnumaa[1])
- N-Y6599 Finland (Eastern Finland Province,[1] Oulu Province,[1] Southern Finland Province[1]), Estonia (Jõgevamaa,[1] Hiiumaa[1]), Russia (Tula Oblast[1])
- N-Y24000
- N-Y16503 Finland (Western Finland Province,[1] Southern Finland Province,[1] Eastern Finland Province,[1] Oulu Province[1]), Sweden (Gävleborg County[1])
- N-Y22106
- N-Y7300
- N-Y19098
- N-BY6010 United States (Virginia[1])
- N-CTS3451
- N-CTS3451* Russian[1]
- N-Y3667
- N-BY33087
- N-BY33087* Finland (Southern Finland Province[1])
- N-BY33088 Finland (Eastern Finland Province[1]), Russia (Novgorod Oblast[1])
- N-CTS657
- N-CTS657* Finland,[1] Russia (Republic of Karelia,[1] Novgorod Oblast[1])
- N-BY70437 Finland (Eastern Finland Province[1])
- N-BY6024 Finland (Eastern Finland Province[1])
- N-Y45925 Russia (Republic of Karelia[1])
- N-Y26750
- N-Y26750* Russia (Arkhangelsk Oblast[1])
- N-A16653 Finland (Eastern Finland Province[1])
- N-PH3568 Finland (Oulu Province,[1] Eastern Finland Province[1])
- N-BY33087
- N-L1022
- N1a1a1a1a1a1-Z4908
- N1a1a1a1a1b-PH1266/Y28526/F4134
- N-Y46746 Russia[1]
- N-Y32732
- N-Y32732* Russia (Saint Petersburg[1])
- N-Y192174
- N-Y32731 Russia (Komi Republic[1]), Sweden[1]
- N1a1a1a1a1c-B479 Nanai,[3] Ulchi[40]
- N1a1a1a1a1a-CTS2929/VL29 Found with high frequency among Lithuanians, Latvians, Estonians, northwestern Russians, Swedish Saami, Karelians, Nenetses, Finns, and Maris, moderate frequency among other Russians, Belarusians, Ukrainians, and Poles, and low frequency among Komis, Mordva, Tatars, Chuvashes, Dolgans, Vepsa, Selkups, Karanogays, and Bashkirs[3]
- N1a1a1a1a2-Z1936,CTS10082 Found with high frequency among Finns, Vepsa, Karelians, Swedish Saami, northwestern Russians, Bashkirs, and Volga Tatars, moderate frequency among other Russians, Komis, Nenetses, Ob-Ugrians, Dolgans, and Siberian Tatars, and low frequency among Mordva, Nganasans, Chuvashes, Estonians, Latvians, Ukrainians, and Karanogays[3]
- N-Y13851
- N-BY199053 France,[59] ancient Western Siberia (Gornaya Bitiya, Iron Age Sargat culture, 300 - 100 BCE[59])
- N-Y13852
- N1a1a1a1a2a1c-PH3340/Y13850
- N1a1a1a1a2a1c1-L1034
- N-Y28538
- N-Y28538* Russia (Khantia-Mansia[1])
- N-L1032 Russia (Khantia-Mansia[1]), Kazakhstan (East Kazakhstan Region[1])
- N-L1442
- N-FT12605
- N-Y23732 Russia (Bashkortostan[1])
- N-Y24222
- N-Y24222* Hungary[1]
- N-Y65329 Russia (Tatarstan Tatar,[1] Bashkortostan[1])
- N-Y62987 Russia (Bashkortostan,[1] Samara Oblast,[1] Tyumen Oblast Siberian Tatar[1])
- N-Y28538
- N1a1a1a1a2a1c2-Y24361 (Y24372)
- N1a1a1a1a2a1c1-L1034
- N1a1a1a1a2a1c-PH3340/Y13850
- N1a1a1a1a2a-Z1934
- N-Y159520
- N-Y159520* Russia (Tambov Oblast[1])
- N-BY184755 Russia (Novgorod Oblast,[1] Voronezh Oblast,[1] Moscow[1])
- N-Y18421
- N-Y18421* Russia (Tambov Oblast[1])
- N-Y19110
- N-Y180247
- N-Y19108
- N-Y19108* Russia (Tatarstan,[1] Kirov Oblast[1])
- N-Y19453
- N-Y19453* Russia (Tatarstan[1])
- N-Y19451 Finland (Western Finland Province[1]), Russia (Republic of Karelia,[1] Tver Oblast[1])
- N-Z1928/CTS2733
- N-YP6091
- N-YP6091* Russia (Irkutsk Oblast[1])
- N-YP6094
- N-Y129131
- N-Y129131* Russia (Kostroma Oblast[1])
- N-A25107 Russia (Kirov Oblast,[1] HGDP Russian[1])
- N-YP6092
- N-YP6092* Russia (Vologda Oblast[1])
- N-B195
- N-B195* Russia (Sverdlovsk Oblast[1])
- N-Y62142 Russia (Republic of Karelia,[1] Arkhangelsk Oblast[1])
- N-Y129131
- N1a1a1a1a2a-Z1925
- N-Z1925* Sweden (Norrbotten County[1])
- N-Y29767 Sweden (Norrbotten County[1])
- N-Y62904 Finland (Western Finland Province,[1] Oulu Province[1])
- N1a1a1a1a2a2a1a1-Z1926
- N-Y20920
- N-Y20920* Finland (Western Finland Province[1]), Norway (Nord-Trøndelag[1])
- N-Y21699 Finland (Western Finland Province,[1] Southern Finland Province[1])
- N-Z1927
- N-Y22108 Finland (Eastern Finland Province,[1] Southern Finland Province,[1] Western Finland Province[1]), Sweden (Stockholm County,[1] Örebro County[1])
- N-Z1933
- N-CTS4329
- N-Y22091
- N-Y22091* Norway (Nordland[1]), Finland (Lapland Province[1])
- N-Y47623
- N-Y31247
- N-Y31247* Russia (Tver Oblast[1])
- N-Y31249
- N-Y90283 Finland (Lapland Province[1])
- N-Y31244 Finland (Western Finland Province,[1] Eastern Finland Province[1])
- N-YP6269
- N-YP6269* Finland (Western Finland Province[1])
- N-Y151660
- N-Y151660* Finland (Eastern Finland Province[1])
- N-BY149208 Sweden (Norrbotten County[1])
- N-CTS8565
- N-BY18114 Finland (Eastern Finland Province[1])
- N-BY30394 Finland (Eastern Finland Province[1]), Russia (Udmurt Republic[1])
- N-Y30513
- N-Y30513* Finland (Southern Finland Province[1])
- N-Y29759
- N-Z4998
- N-Z4998* Finland (Southern Finland Province[1])
- N-FGC65190 Finland (Southern Finland Province, Eastern Finland Province)
- N-Y18414 Finland (Western Finland Province)
- N-Y20910 Finland (Western Finland Province)
- N-BY194138 Finland (Southern Finland Province)
- N-Y28547 Finland (Eastern Finland Province)
- N-BY22141 Finland (Western Finland Province, Eastern Finland Province), United States (Washington)
- N-FT5834 Finland (Western Finland Province, Southern Finland Province)
- N-BY190112 Finland (Western Finland Province, Southern Finland Province)
- N-Y19097 Finland (Eastern Finland Province, Southern Finland Province, Western Finland Province, Oulu Province)
- N-CTS3223 Finland (Southern Finland Province, Eastern Finland Province, Western Finland Province, Oulu Province, Lapland Province), Sweden (Norrbotten County, Västerbotten County, Dalarna County, Värmland County, Skåne County), Russia (Leningrad Oblast)
- N-Y22091
- N-VL62 Russia (Altai Krai,[1] Kostroma Oblast[1]), Estonia (Harjumaa[1]), Finland[1]
- N-VL62* Russia (Chelyabinsk Oblast[1]), Finland[1]
- N-VL60
- N-VL60* Russia (Kurgan Oblast[1])
- N-Y63781 Russia (Vladimir Oblast,[1] Arkhangelsk Oblast[1])
- N-VL72
- N-VL72* Russia (Republic of Karelia[1])
- N-BY30470 Finland (Western Finland Province[1])
- N-Y20915 Finland (Eastern Finland Province[1])
- N-Z1939
- N-Y132561
- N-Z2445
- N-Z35031 Finland (Eastern Finland Province, Western Finland Province), Sweden (Västra Götaland County, Västmanland County)
- N-BY28931
- N-CTS7189
- N-BY142665 Finland (Eastern Finland Province[1])
- N-Y37149 Finland (Eastern Finland Province[1])
- N-Y23568 Finland (Eastern Finland Province[1])
- N-Y80226 Finland (Eastern Finland Province[1])
- N-FT20730 Finland (Eastern Finland Province[1]), Russia (Republic of Karelia[1])
- N-BY62666 Finland (Eastern Finland Province[1]), Norway (Oslo[1]), United States (Virginia,[1] Arkansas[1])
- N-Y24218
- N-BY22038 Finland (Eastern Finland Province,[1] Southern Finland Province,[1] Western Finland Province[1])
- N-Y13974 Finland (Southern Finland Province,[1] Eastern Finland Province,[1] Oulu Province,[1] Western Finland Province[1])
- N-Y135981 Finland (Eastern Finland Province,[1] Oulu Province,[1] Southern Finland Province,[1] Western Finland Province[1]), Norway (Finnmark[1])
- N-Z4747
- N-Y17790 Finland (Eastern Finland Province,[1] Southern Finland Province,[1] Western Finland Province[1])
- N-Z1941
- N-BY22090 Finland (Southern Finland Province,[1] Eastern Finland Province[1])
- N-Y21575 Finland (Western Finland Province,[1] Southern Finland Province,[1] Eastern Finland Province,[1] Oulu Province,[1] Lapland Province[1]), Russia (Leningrad Oblast,[1] Moscow,[1] Tatarstan[1])
- N-Z1940 Finland (Western Finland Province,[1] Southern Finland Province,[1] Eastern Finland Province,[1] Oulu Province,[1] Lapland Province[1]), Russia (Leningrad Oblast[1]), Sweden (Värmland County[1])
- N-CTS4329
- N-Y20920
- N-YP6091
- N-Y159520
- N-Y13851
- N1a1a1a1a3-B197/Y16323
- N1a1a1a1a3a-F4205 Found with high frequency among Buryats[3] and Tsaatans,[28] moderate frequency among Karanogays,[3] Tuvans,[3] Todjins,[28] and Mongols,[3][28] and low frequency among Altaians,[3] Siberian Tatars,[3] Kazakhs,[3] Evenks,[3] Crimean Tatars,[3] Karakalpaks,[3] Uzbeks,[3] and Ukrainians[3]
- N-F4205* China (Inner Mongolia, Gansu, Xinjiang, Henan, etc.[80])
- N-Y16221
- N-Y16221* China (Inner Mongolia[1])
- N-Y16220
- N-Y16220* Ukraine (Luhansk Oblast[1])
- N-F22331 Turkey (Mersin[1]), Poland (Małopolskie[1]), Kazakhstan (Pavlodar Region ca. 1680 ybp[1])
- N-Y63966 Kazakhstan,[1] Turkey,[1] Uzbekistan[1]
- N-Y226011
- N-Z35331 China (Ordos City, Haidong, Chengdu[80])
- N-Y16312/F2288
- N1a1a1a1a3b-B202 Found with high frequency among Chukchis,[3] Koryaks,[3] and Siberian Eskimos[3]
- N1a1a1a1a3a-F4205 Found with high frequency among Buryats[3] and Tsaatans,[28] moderate frequency among Karanogays,[3] Tuvans,[3] Todjins,[28] and Mongols,[3][28] and low frequency among Altaians,[3] Siberian Tatars,[3] Kazakhs,[3] Evenks,[3] Crimean Tatars,[3] Karakalpaks,[3] Uzbeks,[3] and Ukrainians[3]
- N1a1a1a1a1-CTS10760
- N1a1a1a1b-M2019/M2118 Estonia,[1][100] China (Hami, Yan'an, Xi'an, Lüliang, Changzhi, Xilingol League, Chifeng, Cangzhou, Songyuan[80])
- N-M2058 China (Shandong, Liaoning, Henan, Inner Mongolia, Gansu, etc.[80][1])
- N-M2016 China (esp. Xinjiang and Inner Mongolia[80])
- N1a1a1a1b2-A9408 China (Hebei, Shandong, Henan, etc.[80]), Lebanon[53][3]
- N-Y70200 China[1] (Shaanxi, Shanxi, Shandong, etc.[80])
- N-PH1612 China (Yulin, Shangqiu, Dezhou, Tangshan, Shenyang[80])
- N-A9407/A9411 China (Ili Kazakh Autonomous Prefecture[80])
- N-A9416 Hungary (Szabolcs-Szatmár-Bereg[1]), Croatia (Zadar County[1]), Russia (Chuvash Republic[1])
- N-PH1896 Hungary (Borsod-Abaúj-Zemplén[1]), Turkey (Hatay[1][100]), China (Xinjiang, Shandong, Anhui, Heilongjiang, Hebei, Ningxia, Guizhou[80])
- N-A9407/A9411 China (Ili Kazakh Autonomous Prefecture[80])
- N-M2058 China (Shandong, Liaoning, Henan, Inner Mongolia, Gansu, etc.[80][1])
- N1a1a1a1a-L392
- N1a1a1a2-B211 (Y9022) Udmurt,[3] Komi,[3] Chuvash,[3] Ob-Ugrians,[3] Mari,[3] Mordva,[3] Altaian,[3] Belarusian,[3] Karanogay,[3] Karelian,[3] Bashkir,[3] Tatar,[100] Russian,[3][100] Khakas[3]
- N1a1a1a2a-B181
- N-Y182299 Russia (Kirov Oblast, etc.)[1]
- N-Y23788 Russia (Bashkortostan, Komi Republic, etc.)[1]
- N1a1a1a2b-Y23181
- N-Y23183 Russia (Tatarstan, Mordovia, Penza Oblast)[1]
- N-Y143277 Russia (Tatarstan, Bashkortostan)[1]
- N1a1a1a2a-B181
- N1a1a1a1-P298/M2126 China (Urumqi, Kashgar Prefecture, Turpan, Aksu Prefecture, Xianyang, Jincheng, Kaifeng, Qiqihar[80])
- N-Y24317
- N1a1a1a-L708
- N1a1a2-Y23747 Japan (Sado Island[100])
- N1a1a2a-Y23749 Japan[3] (Aichi,[1] Hokkaidō[1])
- N1a1a2b-Y125664
- N-Y125664* Shanghai[1]
- N-MF16376
- N1a1a2b1-MF38607 Anhui,[1] Hunan[1]
- N1a1a2b2-F22150 Hebei,[1] Suzhou[80]
- N-FT281705 China (Shandong, Liaoning, etc.[80])
- N-MF15288/MF15344 China (esp. Jiangsu, but also Henan, Fujian, Shanghai, Heilongjiang,[1] etc.[80]), South Korea[1]
- N-MF55680 China (Hebei, Beijing, Hubei, etc.[80])
- N1a1a1-F1419
- N1a1a-M178
- N-F1360
- N-F1360* China (Shaanxi[1])
- N1a2-F1008/L666
- N-F1101
- N-F1101* China (Shandong[1]), Neolithic Siberia (Trans-Baikal)
- N-F1154
- N-F1154* China (Liaoning[1])
- N-Y23741
- N-Y23741* China[1]
- N1a2a-M128
- N1a2b-B523(P43)
- N1a2b-Y126204, VL67/Z35079, BY29083
- N-VL67/Z35079
- N1a2b1-B478 (P63) Nenets, Nganasans, Dolgans, Tuvans, Todzhins,[28] Khakasses, Shorians, Evens, Altaians, Selkups, Evenks, Mongols (Sart-Kalmak,[28] Torgut,[28] Derbet,[28] Buryat), Yakuts, Ob-Ugrians, Chuvashes
- N1a2b1a-B168 Evens
- N1a2b1b-B169
- N1a2b1b1-B170 Nenets, Khanty-Mansi Autonomous Okrug, Inner Mongolia
- N1a2b1b2-B175 Tuvinians, Mongols, Evenks, Yakuts, Tomsk Oblast
- N1a2b3-B525 Turkey, Tatars, Bashkirs, Kazakhs, Mongols (Xinjiang Kalmyk,[28] Mongolian Torgut[28]), Slovakia, Bulgaria,[1] Ukrainians, Belarusians, Russians
- N-B525* Afghanistan, Arab
- N-BY173686 Russia (Voronezh Oblast[1])
- N-VL83 Turkey[1]
- N-Y136502 Russia (Tatarstan[1]), Bulgaria (Dobrich[1])
- N-Y37153
- N-Y37153* Russia (Tatarstan[1])
- N-BY29131
- N-BY29131* Turkey (Konya[1]), Slovakia (Žilina Region[1])
- N-BY30476 Russia (Tatarstan,[1] Penza Oblast[1])
- N1a2b1-B478 (P63) Nenets, Nganasans, Dolgans, Tuvans, Todzhins,[28] Khakasses, Shorians, Evens, Altaians, Selkups, Evenks, Mongols (Sart-Kalmak,[28] Torgut,[28] Derbet,[28] Buryat), Yakuts, Ob-Ugrians, Chuvashes
- N1a2b2-FGC10872/Y3195
- N1a2b2a-FGC10847/Y3185 (L1419) Vepsas, Maris, Russians (Arkhangelsk Oblast), Komis, Perm Krai, Komi Republic, Ob-Ugrians, Chuvashes, Tatars, Bashkirs, Karelians, Western Finland Province, Tuvans, Buryats, Khakasses, Nganasans, Asian Eskimos
- N1a2b2b-Y23786
- N1a2b2b* Mansis
- N1a2b2b1-B528/Y24384 Udmurts, Komis, Khanties, Tatars, Asian Eskimos, Kirov Oblast, Perm Krai, Medny Island
- N-VL67/Z35079
- N1a2b-Y126204, VL67/Z35079, BY29083
- N-F1101
- N-Z1956
- N1b-F2905
- N1b1-CTS582 China (esp. Jiangsu, Yunnan, Hebei, Shandong, Shanxi, Heilongjiang[80]), Neolithic Shandong
- N1b1a-Y6374/Z8029
- N1b1a1-CTS7324 Beijing[1]
- N1b1a2-L727
- N1b1a2*-L727 Beijing[1]
- N1b1a2a-L732 Belarus[59]
- N1b1a2a1-F839 China (esp. Jiangsu,[80] Shandong,[80] Zhejiang,[80] Shanghai,[1][80] Henan[80])
- N1b1a2a2-Y15972
- N1b1b-Y23789/CTS4309 Iraq[59]
- N1b1a-Y6374/Z8029
- N1b2-M1819/N-M1897/CTS12473/F1173 China,[59] Russian Federation[59]
- N-M1897* ancient DNA from Pingliangtai, Henan (ca. 4,063 [95% CI 3,974 <-> 4,151] ybp)[1]
- N-M1845
- N-M1845* China (Sichuan,[1] Shanghai[1])
- N-M1928
- N-M1928* Sichuan (Han)[1]
- N-Y125475
- N-Y63516 China (Chongqing[1]), Korea (South Pyongan Province,[1] South Chungcheong Province[1])
- N-Y193396 China (Ningxia[1]), Singapore[1]
- N-CTS4714
- N1b1-CTS582 China (esp. Jiangsu, Yunnan, Hebei, Shandong, Shanxi, Heilongjiang[80]), Neolithic Shandong
- N1a-L729
- N2-Y6503
- N2-Y6503* Altai Republic[1]
- N2a-P189.2
- N2a1-Y6516
- N2a1-Y6516*
- N2a1a-Y7310
- N2a1a-Y7310* Romania (Hungarian from Suceava)[1]
- N2a1a1-Y7313
- N2a1a1-Y7313*
- N2a1a1a-BY35494 Russia (Rostov Oblast)[1]
- N2a1a1b-FGC28435
- N2a1a1b-FGC28435* Turkey (Istanbul), Serbia, Montenegro (Plužine), Bosnia and Herzegovina (Republika Srpska), Croatia[1]
- N2a1a1b1-FGC28483 Serbia[1]
- N2a2-Y111068
- N2a2a-FT352925 France (Charente-Maritime[1]), Turkey[1]
- N2a2b-Y101945 United Kingdom (Devon[1]), Russia (Moscow Oblast[1])
- N2a1-Y6516
- N1-Z4762/CTS11499/L735/M2291
- N-M231/Page91, M232/M2188
History of phylogenetic nomenclature
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.[citation needed]
| YCC 2002/2008 (Shorthand) | (α) | (β) | (γ) | (δ) | (ε) | (ζ) | (η) | YCC 2002 (Longhand) | YCC 2005 (Longhand) | YCC 2008 (Longhand) | YCC 2010r (Longhand) | ISOGG 2006 | ISOGG 2007 | ISOGG 2008 | ISOGG 2009 | ISOGG 2010 | ISOGG 2011 | ISOGG 2012 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N-LLY22g | 12 | VIII | 1U | 25 | Eu16 | H5 | F | N* | N | N1 | N1 | - | - | - | - | - | - | - |
| N-M128 | 12 | VIII | 1U | 25 | Eu16 | H5 | F | N1 | N1 | N1a | N1a | - | - | - | - | - | - | - |
| N-P63 | 12 | VIII | 1U | 25 | Eu16 | H5 | F | N2 | N2a | N1b1 | N1b1 | - | - | - | - | - | - | - |
| N-TAT | 12 | VIII | 1I | 26 | Eu13 | H5 | F | N3* | N3 | N1c | N1c | - | - | - | - | - | - | - |
| N-M178 | 16 | VIII | 1I | 26 | Eu14 | H5 | F | N3a* | M178 | N1c1 | N1c1 | - | - | - | - | - | - | - |
| N-P21 | 16 | VIII | 1I | 26 | Eu14 | H5 | F | N3a1 | N3a1 | N1c1a | N1c1a | - | - | - | - | - | - | - |
Sources The following research teams per their publications were represented in the creation of the YCC Tree.
Unreliable mutations (SNPs and UEPs)
The b2/b3 deletion in the AZFc region of the Y-chromosome appears to have occurred independently on at least four different occasions. Therefore, this deletion should not be taken as a unique event polymorphism defining this branch of the Y-chromosome tree (ISOGG 2012).
Links to genetics concepts
- genetic genealogy
- Genetic history of Europe
- Haplogroup
- Haplotype
- Human Y-chromosome DNA haplogroup
- molecular phylogeny
- Paragroup
- Subclade
- Y-chromosome haplogroups in populations of the world
- Y-DNA haplogroups by ethnic group
- Y-DNA haplogroups in populations of East and Southeast Asia
- Y-DNA haplogroups in populations of Europe
Y-DNA N subclades
- N-M231
Y-DNA backbone tree
References
Bibliography
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.