Serratia
Genus of bacteria From Wikipedia, the free encyclopedia
Serratia is a genus of Gram-negative, facultatively anaerobic, rod-shaped bacteria of the family Enterobacteriaceae.[5] They are typically 1–5 μm in length, do not produce spores,[6] and can be found in water, soil, plants, and animals.[7] Some members of this genus produce a characteristic red pigment, prodigiosin, and can be distinguished from other members of the order Enterobacterales by their unique production of three enzymes: DNase (nucA), lipase, and gelatinase (serralysin).[5] Serratia was thought to be a harmless environmental bacteria until it was discovered that the most common species in the genus, S. marcescens, is an opportunistic pathogen of many animals, including humans.[5] In humans, S. marcescens is mostly associated with nosocomial, or hospital-acquired, infections, but can also cause urinary tract infections, pneumonia, and endocarditis.[8] S. marcescens is frequently found in showers, toilet bowls, and around wet tiles as a pinkish to red biofilm but only causes disease in immunocompromised individuals. Aside from S. marcescens, some rare strains of the Serratia species – S. plymuthica, S. liquefaciens, S. rubidaea, and S. odoriferae – have been shown to cause infection such as osteomyelitis and endocarditis.[9]
| Serratia | |
|---|---|
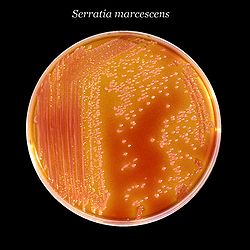 | |
| Serratia marcescens, a typical species, on XLD agar.[1] | |
| Scientific classification | |
| Domain: | Bacteria |
| Kingdom: | Pseudomonadati |
| Phylum: | Pseudomonadota |
| Class: | Gammaproteobacteria |
| Order: | Enterobacterales |
| Family: | Yersiniaceae |
| Genus: | Serratia Bizio, 1823[2][3] |
| Species | |
|
S. aquatilis[4] | |
Habitat
Summarize
Perspective
Various strains of Serratia occupy an eclectic range of habitats: soil, water, plants, insects, and others.[10]
Water
Currently, four species of Serratia have been found in seawater: S. marcescens, S. liquefaciens, S. plymuthica, and S. marinorubra. S. marcescens is the most abundant, comprising approximately half of all strains found.[11] S. aquatilis is a novel species of Serratia found in drinking water.[12]
Plants
The plant types with the highest Serratia prevalence are vegetables, mushrooms, mosses, grasses, and decaying plant material.[13] Serratia has been consistently found in figs and coconuts. S. marcescens and S. ficaria are often found in Calimyrna figs. Several species related to Serratia have also been identified on Smyrna figs and its fig wasps. Only one species of Serratia, S. marinorubra, has been identified on coconuts from various regions of the world, including California, France, and Brazil. Both S. marcescens and certain strains of Enterobacter were also identified in the rhizospheres of coconut palms.[14]
Insects
Serratia is found in over 70 species of healthy, dead, and diseased insects. These include crickets, grasshoppers, bees, aphids, and fruit flies.[10] Most of them reside in insects as bacterial flora and some form mutualistic symbiotic relationships with their hosts. For example, in aphids, strains of S. symbiotica play a key nutritional role by providing the host with vitamins and amino acids.[15]
In 2017 it was reported that Serratia can be genetically engineered to prevent malaria in mosquitos. Research showed 93% fewer Plasmodium parasites than in untreated counterparts.[16][17]
Isolation, identification, and metabolism
Summarize
Perspective
Isolation
S. marcescens is the most characterized species in this genus. During a hot summer in 1819 in Legnaro near Padua, Italy, the town people discovered that their polenta dish turned red.[18] At first, the people believed that this incident was caused by the devil. A pharmacist named Bartolomeo Bizio was appointed to investigate the strange phenomenon.[19] After several experiments, Bizzo presented his results. S. marcescens was first documented as a red-colored putrefaction of polenta by Bartolomeo Bizio in Padua.[19] The bacterium was later named in honor of Italian physicist Serafino Serrati.[19] In 1945, an experiment was designed to establish the pathogenicity of S. marcescens. Captain Tom Paine in the U.S. Army conducted an experiment at Camp Detrick, MD. In this experiment, he exposed four people to the bacteria in an enclosed space.[19] The individuals soon developed symptoms such as body aches, malaise, green sputum production. A few of the individuals developed fever and chills while others still had a fever after 24 hours.[19] Several other experiments were performed throughout the 50s, 60s, and 70s to test the pathogenicity of S. marcescens,[19] but it was not until the 1970s that S. marcescens was confirmed as a human pathogen.
S. liquefaciens is the second best characterized species after S. marcescens. S. liquefaciens was first classified as Aerobacter liquefaciens in the Enterobacter genus by Grimes and Hennerty.[19] The first documentation of S. liquefaciens was in 1971. Over 20 isolates of S. liquefaciens were recovered from different specimens, such as urinary and respiratory. Out of the isolates, six were believed to cause infection in humans.[19] From the 1970s to the 1980s, this species was the cause of several hospital outbreaks. However, the most well-known outbreak occurred in Colorado at a hemodialysis center. During this outbreak, there were 10 S. liquefaciens bloodstream infections.[19]
S. ficaria, a species belonging to the fig tree community, can also be harmful to humans. In 1979, S. ficaria was first isolated from a patient who had a respiratory infection.[19] The organism was isolated from the patient's sputum after she consumed a fig.[19] The organisms continued to be isolated from several humans over the years. The last documented infection caused by S. ficaria was in Greece. A healthy man was bitten by a dog, and the dog bite turned into an abscess. This was the first infection recorded in a healthy individual.
S. fonticola was first found in human specimens in 1985.[19] It is known to cause tissue infections following trauma to the area.[20] The first reported infection caused by S. fonticola was a leg abscess in a woman in France in 1989. In 1991, S. fonticola was the cause of a hand infection in another French woman.[19] S. fonticola has been recovered from several other patients over the years.
There are not many reports of S. quinivoran causing infection in humans. A homeless man in France was admitted to the hospital with a mouth abscess. The man developed pneumonia and respiratory issues. S. quinivoran was recovered from a sample and was later identified as the cause of his organ failure and death.[19] S. rubidaea, S. odorifera, and S. plymuthica are other Serratia species that are human pathogens. However, not all Serratia species are human pathogens. S. entomophia and S. proteamaculans are insect and plant pathogens.
Identification
Species of Serratia have been isolated in a variety of environments, including soil, water, plants, animals and even air. Several methods can be used to study the epidemiology of S. marcescens. Usual enrichment strategies involve the use of media containing antibiotic and antifungal substances. A caprylate-thallous medium seems to be highly preferred for the selective growth of genus Serratia, as it can use caprylic acid as a carbon source.
Serological typing and different types of polymerase chain reaction can be used to identify the Serratia. Biotyping, bacteriocin typing, phage typing, plasmid analysis, and ribotyping can also be used. Most strains of S. marcescens appear red on trypticase soy agar slants when grown at around 25 °C. S. marcescens and S. liquefaciens can be easily confused in the lab when using the analytical profile index system. They can both oxidise arabinose, but only S. liquefaciens can ferment arabinose in peptone water. The virulence of Serratia strains can also be identifiable by type 4 fimbriae, small hair-like projections.[21]
Genome content
The average genome size of most Serratia species has not been documented except for that of S. marcescens, which is 3.57 × 109 daltons. The range of G+C content of each species is as follows: S. marcescens 57.5–60.4%, S. liquefaciens 52.6–54.4%, S. plymuthica 53.3–56.3%, S. marinorubra 53.5–58.5%. S. macescens genome has the highest G+C content among all enterobacteria.[22]
Enzymes and biofilm
Serratia secretes a host of virulence factors, including prodigiosin, biosurfactants, DNAse, lipase, protease, gelatinase, hemolysin, chitinase, chloroperoxidase, and alkaline phosphatase. Prodigiosin, a growth pigment, is often used as a phenotypic identification marker of Serratia species due to its red colorization.[23] Biosurfactants have been isolated from Serratia marcescens, Serratia rubidaea and Serratia faciens for their range of applications, including emulsification, surface, antifouling, antitumor, and antimicrobial activity.[24][25] Endonucleases, such as DNAse, may aid in scavenging activity, allowing them to exploit the environment and maximize the availability of nutrients.[26] Strains producing thermostable lipase,[27] alkaline protease and gelatinase[28] have been isolated from strains causing contact lens-related corneal ulcers in humans. Due to its short half-life and tendency to remain bound to cells upon secretion, hemolysin has scarcely been identified in Serratia. However, some studies employing more accurate detection techniques have evidenced hemolytic activity in almost all strains of Serratia.[29] Plant chitinases are used as defense mechanisms against plant pathogens with which Serratia shares their plant habitat.[30][31] Chloroperoxidase allows the hydrolysis of phosphodiester bonds[32] while alkaline phosphatases are involved in cell signaling processes.
Metabolism
Serratia uses a metabolic enzyme, ADP glucose pyrophosphorylase, with distinct kinetic properties from those found in Enterobacteriaceae in that it is not greatly activated by fructose bisphosphate. ADP glucose pyrophosphorylase from strains of S. marcescens demonstrated optimal activity in buffer at pH 7.5 and 8.0, respectively. It is greatly activated by glycolysis intermediates such as phosphoenolpyruvate, 3-phosphoglycerate, fructose-6-phosphate, and 2-phosphoglycerate.[33]
Pathology
Summarize
Perspective
Most Serratia species are nonpathogenic, but those that are pathogenic typically cause infection in immunocompromised individuals.[34] S. marcescens is the main pathogenic species, infecting animals and plants, but other species that have been reported to infect individuals include Serratia plymuthica, Serratia liquefaciens, Serratia rubidaea, Serratia odorifera, and Serratia fonticola.
Opportunistic human pathogen
S. marcescens is thought to be transmitted through hand-to-hand transmission; in one hospital half of all tested personnels' hands were found to be positive for the pathogen.[35]
Serratia species tend to colonize the respiratory and urinary tracts, rather than the gastrointestinal tract. Serratia infection is responsible for about 2% of nosocomial (hospital-acquired) infections of the bloodstream, lower respiratory tract, urinary tract, surgical wounds, and skin and soft tissues and other ailments that are commonly caused by other bacteria.[9] Outbreaks of S. marcescens meningitis, wound infections, and arthritis have occurred in pediatric wards.[36] Outbreaks of infective endocarditis in IV drug users have been reported.[37]
Cases of Serratia arthritis have been reported in outpatients receiving intra-articular injections.[38]
Opportunistic non-human pathogen
There have been cases of Serratia non-human animal infections. One case of a non-nosocomial infection in animals was found in one study, after S. marcescens was found to be correlated with early abortions in buffalos and cows. The pathogen was isolated in culture after researchers observed reddish vaginal discharge from the cows, and the pathogen was also discovered to be in the semen of a bull, all of which were from the same strain.[39]
Opportunistic plant pathogen
S. marcescens and S. proteamaculans are considered to be opportunistic plant pathogens. S. marcescens causes cucurbit yellow vine disease (CYVD).[40] CYVD was first detected in pumpkin and squash. CYVD infects the phloem tissue in plants and causes wilting, yellowing, phloem discoloration, plant decline, and eventually death.[40] CYVD mainly affects squash, cantaloupe, watermelon, etc. There have been studies that have shown that this disease is transmitted by insects.[40] S. proteamaculans is the only other species known to cause harm to plants. S. proteamaculans is associated with leaf spot disease. Leaf spot disease is usually caused by a fungus, but can also be caused by bacteria (e.g. S. proteamaculans). Leaf spot disease appears as brown or dark spots on leaves and can permanently damage plants. The sizes and colors of these spots can vary.
See also
- Delftia tsuruhatensis – a bacterium that naturally prevents malaria.
- Wolbachia – a genus of bacteria that can be used to control dengue.
References
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
