Ligand-gated ion channel
Type of ion channel transmembrane protein From Wikipedia, the free encyclopedia
Ligand-gated ion channels (LICs, LGIC), also commonly referred to as ionotropic receptors, are a group of transmembrane ion-channel proteins which open to allow ions such as Na+, K+, Ca2+, and/or Cl− to pass through the membrane in response to the binding of a chemical messenger (i.e. a ligand), such as a neurotransmitter.[1][2][3]
| Neurotransmitter-gated ion-channel transmembrane region | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Ligand-gated ion channel | |||||||||
| Identifiers | |||||||||
| Symbol | Neur_chan_memb | ||||||||
| Pfam | PF02932 | ||||||||
| InterPro | IPR006029 | ||||||||
| PROSITE | PDOC00209 | ||||||||
| SCOP2 | 1cek / SCOPe / SUPFAM | ||||||||
| TCDB | 1.A.9 | ||||||||
| OPM superfamily | 14 | ||||||||
| OPM protein | 2bg9 | ||||||||
| |||||||||

- Ion-channel-linked receptor
- Ions
- Ligand (such as acetylcholine)
When a presynaptic neuron is excited, it releases a neurotransmitter from vesicles into the synaptic cleft. The neurotransmitter then binds to receptors located on the postsynaptic neuron. If these receptors are ligand-gated ion channels, a resulting conformational change opens the ion channels, which leads to a flow of ions across the cell membrane. This, in turn, results in either a depolarization, for an excitatory receptor response, or a hyperpolarization, for an inhibitory response.
These receptor proteins are typically composed of at least two different domains: a transmembrane domain which includes the ion pore, and an extracellular domain which includes the ligand binding location (an allosteric binding site). This modularity has enabled a 'divide and conquer' approach to finding the structure of the proteins (crystallising each domain separately). The function of such receptors located at synapses is to convert the chemical signal of presynaptically released neurotransmitter directly and very quickly into a postsynaptic electrical signal. Many LICs are additionally modulated by allosteric ligands, by channel blockers, ions, or the membrane potential. LICs are classified into three superfamilies which lack evolutionary relationship: cys-loop receptors, ionotropic glutamate receptors and ATP-gated channels.
Cys-loop receptors
Summarize
Perspective

The cys-loop receptors are named after a characteristic loop formed by a disulfide bond between two cysteine residues in the N terminal extracellular domain. They are part of a larger family of pentameric ligand-gated ion channels that usually lack this disulfide bond, hence the tentative name "Pro-loop receptors".[4][5] A binding site in the extracellular N-terminal ligand-binding domain gives them receptor specificity for (1) acetylcholine (AcCh), (2) serotonin, (3) glycine, (4) glutamate and (5) γ-aminobutyric acid (GABA) in vertebrates. The receptors are subdivided with respect to the type of ion that they conduct (anionic or cationic) and further into families defined by the endogenous ligand. They are usually pentameric with each subunit containing 4 transmembrane helices constituting the transmembrane domain, and a beta sheet sandwich type, extracellular, N terminal, ligand binding domain.[6] Some also contain an intracellular domain like shown in the image.
The prototypic ligand-gated ion channel is the nicotinic acetylcholine receptor. It consists of a pentamer of protein subunits (typically ααβγδ), with two binding sites for acetylcholine (one at the interface of each alpha subunit). When the acetylcholine binds it alters the receptor's configuration (twists the T2 helices which moves the leucine residues, which block the pore, out of the channel pathway) and causes the constriction in the pore of approximately 3 angstroms to widen to approximately 8 angstroms so that ions can pass through. This pore allows Na+ ions to flow down their electrochemical gradient into the cell. With a sufficient number of channels opening at once, the inward flow of positive charges carried by Na+ ions depolarizes the postsynaptic membrane sufficiently to initiate an action potential.
A bacterial homologue to an LIC has been identified, hypothesized to act nonetheless as a chemoreceptor.[4] This prokaryotic nAChR variant is known as the GLIC receptor, after the species in which it was identified; Gloeobacter Ligand-gated Ion Channel.
Structure
Cys-loop receptors have structural elements that are well conserved, with a large extracellular domain (ECD) harboring an alpha-helix and 10 beta-strands. Following the ECD, four transmembrane segments (TMSs) are connected by intracellular and extracellular loop structures.[7] Except the TMS 3-4 loop, their lengths are only 7-14 residues. The TMS 3-4 loop forms the largest part of the intracellular domain (ICD) and exhibits the most variable region between all of these homologous receptors. The ICD is defined by the TMS 3-4 loop together with the TMS 1-2 loop preceding the ion channel pore.[7] Crystallization has revealed structures for some members of the family, but to allow crystallization, the intracellular loop was usually replaced by a short linker present in prokaryotic cys-loop receptors, so their structures as not known. Nevertheless, this intracellular loop appears to function in desensitization, modulation of channel physiology by pharmacological substances, and posttranslational modifications. Motifs important for trafficking are therein, and the ICD interacts with scaffold proteins enabling inhibitory synapse formation.[7]
Cationic cys-loop receptors
| Type | Class | IUPHAR-recommended protein name [8] |
Gene | Previous names |
|---|---|---|---|---|
| Serotonin (5-HT) |
5-HT3 | 5-HT3A 5-HT3B 5-HT3C 5-HT3D 5-HT3E |
HTR3A HTR3B HTR3C HTR3D HTR3E |
5-HT3A 5-HT3B 5-HT3C 5-HT3D 5-HT3E |
| Nicotinic acetylcholine (nAChR) |
alpha | α1 α2 α3 α4 α5 α6 α7 α9 α10 |
CHRNA1 CHRNA2 CHRNA3 CHRNA4 CHRNA5 CHRNA6 CHRNA7 CHRNA9 CHRNA10 |
ACHRA, ACHRD, CHRNA, CMS2A, FCCMS, SCCMS |
| beta | β1 β2 β3 β4 |
CHRNB1 CHRNB2 CHRNB3 CHRNB4 |
CMS2A, SCCMS, ACHRB, CHRNB, CMS1D EFNL3, nAChRB2 | |
| gamma | γ | CHRNG | ACHRG | |
| delta | δ | CHRND | ACHRD, CMS2A, FCCMS, SCCMS | |
| epsilon | ε | CHRNE | ACHRE, CMS1D, CMS1E, CMS2A, FCCMS, SCCMS | |
| Zinc-activated ion channel (ZAC) |
ZAC | ZACN | ZAC1, L2m LICZ, LICZ1 |
Anionic cys-loop receptors
| Type | Class | IUPHAR-recommended protein name[8] |
Gene | Previous names |
|---|---|---|---|---|
| GABAA | alpha | α1 α2 α3 α4 α5 α6 |
GABRA1 GABRA2 GABRA3 GABRA4 GABRA5 GABRA6 |
EJM, ECA4 |
| beta | β1 β2 β3 |
GABRB1 GABRB2 GABRB3 |
ECA5 | |
| gamma | γ1 γ2 γ3 |
GABRG1 GABRG2 GABRG3 |
CAE2, ECA2, GEFSP3 | |
| delta | δ | GABRD | ||
| epsilon | ε | GABRE | ||
| pi | π | GABRP | ||
| theta | θ | GABRQ | ||
| rho | ρ1 ρ2 ρ3 |
GABRR1 GABRR2 GABRR3 |
GABAC[9] | |
| Glycine (GlyR) |
alpha | α1 α2 α3 α4 |
GLRA1 GLRA2 GLRA3 GLRA4 |
STHE |
| beta | β | GLRB |
Ionotropic glutamate receptors
Summarize
Perspective
The ionotropic glutamate receptors bind the neurotransmitter glutamate. They form tetramers, with each subunit consisting of an extracellular amino terminal domain (ATD, which is involved tetramer assembly), an extracellular ligand binding domain (LBD, which binds glutamate), and a transmembrane domain (TMD, which forms the ion channel). The transmembrane domain of each subunit contains three transmembrane helices as well as a half membrane helix with a reentrant loop. The structure of the protein starts with the ATD at the N terminus followed by the first half of the LBD which is interrupted by helices 1,2 and 3 of the TMD before continuing with the final half of the LBD and then finishing with helix 4 of the TMD at the C terminus. This means there are three links between the TMD and the extracellular domains. Each subunit of the tetramer has a binding site for glutamate formed by the two LBD sections forming a clamshell like shape. Only two of these sites in the tetramer need to be occupied to open the ion channel. The pore is mainly formed by the half helix 2 in a way which resembles an inverted potassium channel.

| Type | Class | IUPHAR-recommended protein name [8] |
Gene | Previous names |
|---|---|---|---|---|
| AMPA | GluA | GluA1 GluA2 GluA3 GluA4 |
GRIA1 GRIA2 GRIA3 GRIA4 |
GLUA1, GluR1, GluRA, GluR-A, GluR-K1, HBGR1 GLUA2, GluR2, GluRB, GluR-B, GluR-K2, HBGR2 GLUA3, GluR3, GluRC, GluR-C, GluR-K3 GLUA4, GluR4, GluRD, GluR-D |
| Kainate | GluK | GluK1 GluK2 GluK3 GluK4 GluK5 |
GRIK1 GRIK2 GRIK3 GRIK4 GRIK5 |
GLUK5, GluR5, GluR-5, EAA3 GLUK6, GluR6, GluR-6, EAA4 GLUK7, GluR7, GluR-7, EAA5 GLUK1, KA1, KA-1, EAA1 GLUK2, KA2, KA-2, EAA2 |
| NMDA | GluN | GluN1 NRL1A NRL1B |
GRIN1 GRINL1A GRINL1B |
GLUN1, NMDA-R1, NR1, GluRξ1 |
| GluN2A GluN2B GluN2C GluN2D |
GRIN2A GRIN2B GRIN2C GRIN2D |
GLUN2A, NMDA-R2A, NR2A, GluRε1 GLUN2B, NMDA-R2B, NR2B, hNR3, GluRε2 GLUN2C, NMDA-R2C, NR2C, GluRε3 GLUN2D, NMDA-R2D, NR2D, GluRε4 | ||
| GluN3A GluN3B |
GRIN3A GRIN3B |
GLUN3A, NMDA-R3A, NMDAR-L, chi-1 GLU3B, NMDA-R3B | ||
| ‘Orphan’ | (GluD) | GluD1 GluD2 |
GRID1 GRID2 |
GluRδ1 GluRδ2 |
AMPA receptor

The α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (also known as AMPA receptor, or quisqualate receptor) is a non-NMDA-type ionotropic transmembrane receptor for glutamate that mediates fast synaptic transmission in the central nervous system (CNS). Its name is derived from its ability to be activated by the artificial glutamate analog AMPA. The receptor was first named the "quisqualate receptor" by Watkins and colleagues after a naturally occurring agonist quisqualate and was only later given the label "AMPA receptor" after the selective agonist developed by Tage Honore and colleagues at the Royal Danish School of Pharmacy in Copenhagen.[10] AMPARs are found in many parts of the brain and are the most commonly found receptor in the nervous system. The AMPA receptor GluA2 (GluR2) tetramer was the first glutamate receptor ion channel to be crystallized. Ligands include:
- Agonists: Glutamate, AMPA, 5-Fluorowillardiine, Domoic acid, Quisqualic acid, etc.
- Antagonists: CNQX, Kynurenic acid, NBQX, Perampanel, Piracetam, etc.
- Positive allosteric modulators: Aniracetam, Cyclothiazide, CX-516, CX-614, etc.
- Negative allosteric modulators: Ethanol, Perampanel, Talampanel, GYKI-52,466, etc.
NMDA receptors
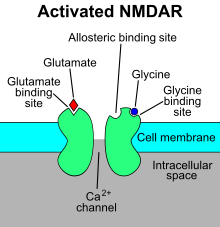
The N-methyl-D-aspartate receptor (NMDA receptor) – a type of ionotropic glutamate receptor – is a ligand-gated ion channel that is gated by the simultaneous binding of glutamate and a co-agonist (i.e., either D-serine or glycine).[11] Studies show that the NMDA receptor is involved in regulating synaptic plasticity and memory.[12][13]
The name "NMDA receptor" is derived from the ligand N-methyl-D-aspartate (NMDA), which acts as a selective agonist at these receptors. When the NMDA receptor is activated by the binding of two co-agonists, the cation channel opens, allowing Na+ and Ca2+ to flow into the cell, in turn raising the cell's electric potential. Thus, the NMDA receptor is an excitatory receptor. At resting potentials, the binding of Mg2+ or Zn2+ at their extracellular binding sites on the receptor blocks ion flux through the NMDA receptor channel. "However, when neurons are depolarized, for example, by intense activation of colocalized postsynaptic AMPA receptors, the voltage-dependent block by Mg2+ is partially relieved, allowing ion influx through activated NMDA receptors. The resulting Ca2+ influx can trigger a variety of intracellular signaling cascades, which can ultimately change neuronal function through activation of various kinases and phosphatases".[14] Ligands include:
- Primary endogenous co-agonists: glutamate and either D-serine or glycine
- Other agonists : aminocyclopropanecarboxylic acid; D-cycloserine; L-aspartate; quinolinate, etc.
- Partial agonists : N-methyl-D-aspartic acid (NMDA); NRX-1074; 3,5-dibromo-L-phenylalanine,[15] etc.
- Antagonists: ketamine, PCP, dextropropoxyphene, ketobemidone, tramadol, kynurenic acid (endogenous), etc.
ATP-gated channels

ATP-gated channels open in response to binding the nucleotide ATP. They form trimers with two transmembrane helices per subunit and both the C and N termini on the intracellular side.
Clinical relevance
Ligand-gated ion channels are likely to be the major site at which anaesthetic agents and ethanol have their effects, although unequivocal evidence of this is yet to be established.[16][17] In particular, the GABA and NMDA receptors are affected by anaesthetic agents at concentrations similar to those used in clinical anaesthesia.[18]
By understanding the mechanism and exploring the chemical/biological/physical component that could function on those receptors, more and more clinical applications are proven by preliminary experiments or FDA. Memantine is approved by the U.S. F.D.A and the European Medicines Agency for the treatment of moderate-to-severe Alzheimer's disease,[19] and has now received a limited recommendation by the UK's National Institute for Health and Care Excellence for patients who fail other treatment options.[20] Agomelatine, is a type of drug that acts on a dual melatonergic-serotonergic pathway, which have shown its efficacy in the treatment of anxious depression during clinical trials,[21][22] study also suggests the efficacy in the treatment of atypical and melancholic depression.[23]
See also
References
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
