EF-Tu
Prokaryotic elongation factor From Wikipedia, the free encyclopedia
EF-Tu (elongation factor thermo unstable) is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes.[2][3][4] It is found in eukaryotic mitochondria as TUFM.[5]
| Elongation Factor Thermo Unstable | |||||||||
|---|---|---|---|---|---|---|---|---|---|
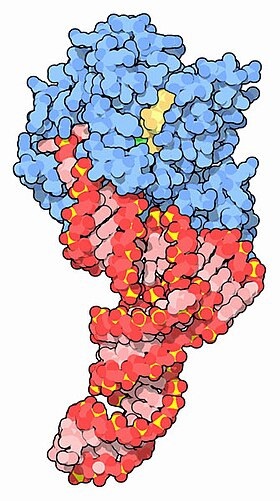 EF-Tu (blue) complexed with tRNA (red) and GTP (yellow) [1] | |||||||||
| Identifiers | |||||||||
| Symbol | EF-Tu | ||||||||
| Pfam | GTP_EFTU | ||||||||
| Pfam clan | CL0023 | ||||||||
| InterPro | IPR004541 | ||||||||
| PROSITE | PDOC00273 | ||||||||
| CATH | 1ETU | ||||||||
| SCOP2 | 1ETU / SCOPe / SUPFAM | ||||||||
| CDD | cd00881 | ||||||||
| |||||||||
As a family of elongation factors, EF-Tu also includes its eukaryotic and archaeal homolog, the alpha subunit of eEF-1 (EF-1A).
Background
Summarize
Perspective
Elongation factors are part of the mechanism that synthesizes new proteins through translation in the ribosome. Transfer RNAs (tRNAs) carry the individual amino acids that become integrated into a protein sequence, and have an anticodon for the specific amino acid that they are charged with. Messenger RNA (mRNA) carries the genetic information that encodes the primary structure of a protein, and contains codons that code for each amino acid. The ribosome creates the protein chain by following the mRNA code and integrating the amino acid of an aminoacyl-tRNA (also known as a charged tRNA) to the growing polypeptide chain.[6][7]
There are three sites on the ribosome for tRNA binding. These are the aminoacyl/acceptor site (abbreviated A), the peptidyl site (abbreviated P), and the exit site (abbreviated E). The P-site holds the tRNA connected to the polypeptide chain being synthesized, and the A-site is the binding site for a charged tRNA with an anticodon complementary to the mRNA codon associated with the site. After binding of a charged tRNA to the A-site, a peptide bond is formed between the growing polypeptide chain on the P-site tRNA and the amino acid of the A-site tRNA, and the entire polypeptide is transferred from the P-site tRNA to the A-site tRNA. Then, in a process catalyzed by the prokaryotic elongation factor EF-G (historically known as translocase), the coordinated translocation of the tRNAs and mRNA occurs, with the P-site tRNA moving to the E-site, where it dissociates from the ribosome, and the A-site tRNA moves to take its place in the P-site.[6][7]
Biological functions
Summarize
Perspective

Protein synthesis
EF-Tu participates in the polypeptide elongation process of protein synthesis. In prokaryotes, the primary function of EF-Tu is to transport the correct aa-tRNA to the A-site of the ribosome. As a G-protein, it uses GTP to facilitate its function. Outside of the ribosome, EF-Tu complexed with GTP (EF-Tu • GTP) complexes with aa-tRNA to form a stable EF-Tu • GTP • aa-tRNA ternary complex.[8] EF-Tu • GTP binds all correctly-charged aa-tRNAs with approximately identical affinity, except those charged with initiation residues and selenocysteine.[9][10] This can be accomplished because although different amino acid residues have varying side-chain properties, the tRNAs associated with those residues have varying structures to compensate for differences in side-chain binding affinities.[11][12]
The binding of an aa-tRNA to EF-Tu • GTP allows for the ternary complex to be translocated to the A-site of an active ribosome, in which the anticodon of the tRNA binds to the codon of the mRNA. If the correct anticodon binds to the mRNA codon, the ribosome changes configuration and alters the geometry of the GTPase domain of EF-Tu, resulting in the hydrolysis of the GTP associated with the EF-Tu to GDP and Pi. As such, the ribosome functions as a GTPase-activating protein (GAP) for EF-Tu. Upon GTP hydrolysis, the conformation of EF-Tu changes drastically and dissociates from the aa-tRNA and ribosome complex.[4][13] The aa-tRNA then fully enters the A-site, where its amino acid is brought near the P-site's polypeptide and the ribosome catalyzes the covalent transfer of the polypeptide onto the amino acid.[10]
In the cytoplasm, the deactivated EF-Tu • GDP is acted on by the prokaryotic elongation factor EF-Ts, which causes EF-Tu to release its bound GDP. Upon dissociation of EF-Ts, EF-Tu is able to complex with a GTP due to the 5– to 10–fold higher concentration of GTP than GDP in the cytoplasm, resulting in reactivated EF-Tu • GTP, which can then associate with another aa-tRNA.[8][13]
Maintaining translational accuracy
EF-Tu contributes to translational accuracy in three ways. In translation, a fundamental problem is that near-cognate anticodons have similar binding affinity to a codon as cognate anticodons, such that anticodon-codon binding in the ribosome alone is not sufficient to maintain high translational fidelity. This is addressed by the ribosome not activating the GTPase activity of EF-Tu if the tRNA in the ribosome's A-site does not match the mRNA codon, thus preferentially increasing the likelihood for the incorrect tRNA to leave the ribosome.[14] Additionally, regardless of tRNA matching, EF-Tu also induces a delay after freeing itself from the aa-tRNA, before the aa-tRNA fully enters the A-site (a process called accommodation). This delay period is a second opportunity for incorrectly charged aa-tRNAs to move out of the A-site before the incorrect amino acid is irreversibly added to the polypeptide chain.[15][16] A third mechanism is the less well understood function of EF-Tu to crudely check aa-tRNA associations and reject complexes where the amino acid is not bound to the correct tRNA coding for it.[11]
Other functions
EF-Tu has been found in large quantities in the cytoskeletons of bacteria, co-localizing underneath the cell membrane with MreB, a cytoskeletal element that maintains cell shape.[17][18] Defects in EF-Tu have been shown to result in defects in bacterial morphology.[19] Additionally, EF-Tu has displayed some chaperone-like characteristics, with some experimental evidence suggesting that it promotes the refolding of a number of denatured proteins in vitro.[20][21] EF-Tu has been found to moonlight on the cell surface of the pathogenic bacteria Staphylococcus aureus, Mycoplasma pneumoniae, and Mycoplasma hyopneumoniae, where EF-Tu is processed and can bind to a range of host molecules.[22] In Bacillus cereus, EF-Tu also moonlights on the surface, where it acts as an environmental sensor and binds to substance P.[23]
Structure
Summarize
Perspective

EF-Tu is a monomeric protein with molecular weight around 43 kDa in Escherichia coli.[24][25][26] The protein consists of three structural domains: a GTP-binding domain and two oligonucleotide-binding domains, often referred to as domain 2 and domain 3. The N-terminal domain I of EF-Tu is the GTP-binding domain. It consists of a six beta-strand core flanked by six alpha-helices.[8] Domains II and III of EF-Tu, the oligonucleotide-binding domains, both adopt beta-barrel structures.[27][28]
The GTP-binding domain I undergoes a dramatic conformational change upon GTP hydrolysis to GDP, allowing EF-Tu to dissociate from aa-tRNA and leave the ribosome.[29] Reactivation of EF-Tu is achieved by GTP binding in the cytoplasm, which leads to a significant conformational change that reactivates the tRNA-binding site of EF-Tu. In particular, GTP binding to EF-Tu results in a ~90° rotation of domain I relative to domains II and III, exposing the residues of the tRNA-binding active site.[30]
Domain 2 adopts a beta-barrel structure, and is involved in binding to charged tRNA.[31] This domain is structurally related to the C-terminal domain of EF2, to which it displays weak sequence similarity. This domain is also found in other proteins such as translation initiation factor IF-2 and tetracycline-resistance proteins. Domain 3 represents the C-terminal domain, which adopts a beta-barrel structure, and is involved in binding to both charged tRNA and to EF1B (or EF-Ts).[32]
Evolution
This section may require cleanup to meet Wikipedia's quality standards. The specific problem is: Still terrible, consider just transcluding the section in "further". (December 2023) |
The GTP-binding domain is conserved in both EF-1alpha/EF-Tu and also in EF-2/EF-G and thus seems typical for GTP-dependent proteins which bind non-initiator tRNAs to the ribosome. The GTP-binding translation factor family also includes the eukaryotic peptide chain release factor GTP-binding subunits[33] and prokaryotic peptide chain release factor 3 (RF-3);[34] the prokaryotic GTP-binding protein lepA and its homologue in yeast (GUF1) and Caenorhabditis elegans (ZK1236.1); yeast HBS1;[35] rat Eef1a1 (formerly "statin S1");[36] and the prokaryotic selenocysteine-specific elongation factor selB.[37]
Disease relevance
Along with the ribosome, EF-Tu is one of the most important targets for antibiotic-mediated inhibition of translation.[8] Antibiotics targeting EF-Tu can be categorized into one of two groups, depending on the mechanism of action, and one of four structural families. The first group includes the antibiotics pulvomycin and GE2270A, and inhibits the formation of the ternary complex.[38] The second group includes the antibiotics kirromycin and enacyloxin, and prevents the release of EF-Tu from the ribosome after GTP hydrolysis.[39][40][41]
See also
- Prokaryotic elongation factors
- EF-Ts (elongation factor thermo stable)
- EF-G (elongation factor G)
- EF-P (elongation factor P)
- eEF-1
- EFR (EF-Tu receptor)
References
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
