Catalytic triad
Set of three coordinated amino acids From Wikipedia, the free encyclopedia
A catalytic triad is a set of three coordinated amino acid residues that can be found in the active site of some enzymes.[1][2] Catalytic triads are most commonly found in hydrolase and transferase enzymes (e.g. proteases, amidases, esterases, acylases, lipases and β-lactamases). An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence (primary structure).[3]

As well as divergent evolution of function (and even the triad's nucleophile), catalytic triads show some of the best examples of convergent evolution. Chemical constraints on catalysis have led to the same catalytic solution independently evolving in at least 23 separate superfamilies.[2] Their mechanism of action is consequently one of the best studied in biochemistry.[4][5]
History
Summarize
Perspective
In the 1950s, a serine residue was identified as the catalytic nucleophile of trypsin and chymotrypsin (first purified in the 1930s)[6] by diisopropyl fluorophosphate modification.[7] The structure of chymotrypsin was solved by X-ray crystallography in the 1960s, showing the orientation of the catalytic triad in the active site.[8] Other proteases were sequenced and aligned to reveal a family of related proteases,[9][10][11] now called the S1 family. Simultaneously, the structures of the evolutionarily unrelated papain and subtilisin proteases were found to contain analogous triads. The 'charge-relay' mechanism for the activation of the nucleophile by the other triad members was proposed in the late 1960s.[12] As more protease structures were solved by X-ray crystallography in the 1970s and 80s, homologous (such as TEV protease) and analogous (such as papain) triads were found.[13][14][15] The MEROPS classification system in the 1990s and 2000s began classing proteases into structurally related enzyme superfamilies and so acts as a database of the convergent evolution of triads in over 20 superfamilies.[16][17] Understanding how chemical constraints on evolution led to the convergence of so many enzyme families on the same triad geometries has developed in the 2010s.[2]
Researchers have since conducted increasingly detailed investigations of the triad's exact catalytic mechanism. Of particular contention in the 1990s and 2000s was whether low-barrier hydrogen bonding contributed to catalysis,[18][19][20] or whether ordinary hydrogen bonding is sufficient to explain the mechanism.[21][22] The massive body of work on the charge-relay, covalent catalysis used by catalytic triads has led to the mechanism being the best characterised in all of biochemistry.[4][5][21]
Function
Summarize
Perspective
Enzymes that contain a catalytic triad use it for one of two reaction types: either to split a substrate (hydrolases) or to transfer one portion of a substrate over to a second substrate (transferases). Triads are an inter-dependent set of residues in the active site of an enzyme and act in concert with other residues (e.g. binding site and oxyanion hole) to achieve nucleophilic catalysis. These triad residues act together to make the nucleophile member highly reactive, generating a covalent intermediate with the substrate that is then resolved to complete catalysis.[23]
Mechanism
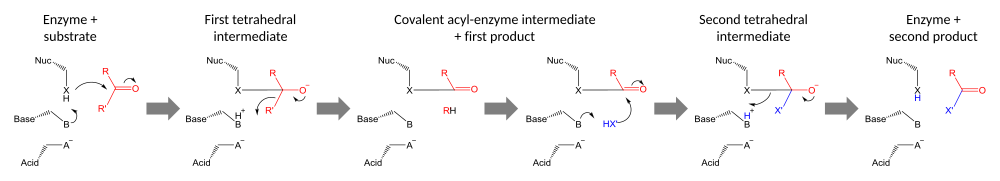
Catalytic triads perform covalent catalysis using a residue as a nucleophile. The reactivity of the nucleophilic residue is increased by the functional groups of the other triad members. The nucleophile is polarised and oriented by the base, which is itself bound and stabilised by the acid.[24]
Catalysis is performed in two stages. First, the activated nucleophile attacks the carbonyl carbon and forces the carbonyl oxygen to accept an electron pair, leading to a tetrahedral intermediate. The resulting build-up of negative charge is typically stabilized by an oxyanion hole within the active site. The intermediate then collapses back to a carbonyl, ejecting the first half of the substrate, but leaving the second half still covalently bound to the enzyme as an acyl-enzyme intermediate. Although general-acid catalysis for breakdown of the First and Second tetrahedral intermediate may occur by the path shown in the diagram, evidence supporting such a mechanism with chymotrypsin[25] has been controverted.[26]
The second stage of catalysis is the resolution of the acyl-enzyme intermediate by the attack of a second substrate. If the substrate is water then hydrolysis results; if it is an organic molecule then that molecule is transferred onto the first substrate. Attack by the second substrate forms a new tetrahedral intermediate, which resolves by ejecting the enzyme's nucleophile, releasing the second product and regenerating free enzyme.[27]
Identity of triad members
Summarize
Perspective

Nucleophile
The side-chain of the nucleophilic residue performs covalent catalysis on the substrate. The lone pair of electrons present on the oxygen or sulfur attacks the electropositive carbonyl carbon.[3] The 20 naturally occurring biological amino acids do not contain any sufficiently nucleophilic functional groups for many difficult catalytic reactions. Embedding the nucleophile in a triad increases its reactivity for efficient catalysis. The most commonly used nucleophiles are the hydroxyl (OH) of serine and the thiol/thiolate ion (SH/S−) of cysteine.[2] Alternatively, threonine proteases use the secondary hydroxyl of threonine, however due to steric hindrance of the side chain's extra methyl group, such proteases use their N-terminal amide as the base rather than a separate amino acid.[1][28]
Use of oxygen or sulfur as the nucleophilic atom causes minor differences in catalysis. Compared to oxygen, sulfur's extra d orbital makes it larger (by 0.4 Å)[29] and softer, allows it to form longer bonds (dC-X and dX-H by 1.3-fold), and gives it a lower pKa (by 5 units).[30] Serine is therefore more dependent than cysteine on optimal orientation of the acid-base triad members to reduce its pKa[30] in order to achieve concerted deprotonation with catalysis.[2] The low pKa of cysteine works to its disadvantage in the resolution of the first tetrahedral intermediate as unproductive reversal of the original nucleophilic attack is the more favourable breakdown product.[2] The triad base is therefore preferentially oriented to protonate the leaving group amide to ensure that it is ejected to leave the enzyme sulfur covalently bound to the substrate N-terminus. Finally, resolution of the acyl-enzyme (to release the substrate C-terminus) requires serine to be re-protonated whereas cysteine can leave as S−. Sterically, the sulfur of cysteine also forms longer bonds and has a bulkier van der Waals radius[2] and if mutated to serine can be trapped in unproductive orientations in the active site.[29]
Very rarely, the selenium atom of the uncommon amino acid selenocysteine is used as a nucleophile.[31] The deprotonated Se− state is strongly favoured when in a catalytic triad.[31]
Base
Since no natural amino acids are strongly nucleophilic, the base in a catalytic triad polarises and deprotonates the nucleophile to increase its reactivity.[3] Additionally, it protonates the first product to aid leaving group departure.[32]
The base is most commonly histidine since its pKa allows for effective base catalysis, hydrogen bonding to the acid residue, and deprotonation of the nucleophile residue.[1] β-lactamases such as TEM-1 use a lysine residue as the base. Because lysine's pKa is so high (pKa=11), a glutamate and several other residues act as the acid to stabilise its deprotonated state during the catalytic cycle.[33][34] Threonine proteases use their N-terminal amide as the base, since steric crowding by the catalytic threonine's methyl prevents other residues from being close enough.[35][36]
Acid
The acidic triad member forms a hydrogen bond with the basic residue, leading to mutual alignment via restriction of the basic residue's side-chain rotation. The positive charge on the basic residue is simultaneously stabilised, leading to its polarisation.[3] Two amino acids have acidic side chains at physiological pH (aspartate or glutamate) and so are the most common members of the acidic triad residue.[3] Cytomegalovirus protease[b] uses a pair of histidines, one as the base, as usual, and one as the acid.[1] The second histidine is not as effective an acid as the more common aspartate or glutamate, leading to a lower catalytic efficiency.[37]
Examples of triads
Summarize
Perspective

Ser-His-Asp
The Serine-Histidine-Aspartate motif is one of the most thoroughly characterised catalytic motifs in biochemistry.[3] The triad is exemplified by chymotrypsin,[c] a model serine protease from the PA superfamily which uses its triad to hydrolyse protein backbones. The aspartate is hydrogen bonded to the histidine, increasing the pKa of its imidazole nitrogen from 7 to around 12. Histidine is thus able to act as a powerful general base, activating the serine nucleophile. The histidine base aids the first leaving group by donating a proton, and also activates the hydrolytic water substrate by abstracting a proton as the remaining OH− attacks the acyl-enzyme intermediate.[23]
The same triad has also convergently evolved in α/β hydrolases such as some lipases and esterases, however orientation of the triad members is reversed.[38][39] Additionally, brain acetyl hydrolase (which has the same fold as a small G-protein) has also been found to have this triad.[40]
Cys-His-Asp
The second most studied triad is the Cysteine-Histidine-Aspartate motif.[2] Several families of cysteine proteases use this triad set, for example TEV protease[a] and papain.[d] The triad acts similarly to serine protease triads, with a few notable differences. Due to cysteine's low pKa, the importance of the Asp to catalysis varies and several cysteine proteases are effectively Cys-His dyads (e.g. hepatitis A virus protease), whilst in others the cysteine is already deprotonated before catalysis begins (e.g. papain).[41] This triad is also used by some amidases, such as N-glycanase to hydrolyse non-peptide C-N bonds.[42]
Ser-His-His
The triad of cytomegalovirus protease[b] uses histidine as both the acid and base triad members. Removing the acid histidine results in only a 10-fold activity loss (compared to >10,000-fold when aspartate is removed from chymotrypsin). This triad has been interpreted as a possible way of generating a less active enzyme to control cleavage rate.[28]
Ser-Glu-Asp
An unusual triad is found in sedolisin proteases.[e] The low pKa of the glutamate carboxylate group means that it only acts as a base in the triad at very low pH. The triad is hypothesised to be an adaptation to specific environments like acidic hot springs (e.g. kumamolysin) or cell lysosome (e.g. tripeptidyl peptidase).[28]
Cys-His-Ser
The endothelial protease vasohibin[f] uses a cysteine as the nucleophile, but a serine to coordinate the histidine base.[43][44] Despite the serine being a poor acid, it is still effective in orienting the histidine in the catalytic triad.[43] Some homologues alternatively have a threonine instead of serine at the acid location.[43]
Thr-Nter, Ser-Nter and Cys-Nter
Threonine proteases, such as the proteasome protease subunit[g] and ornithine acyltransferases[h] use the secondary hydroxyl of threonine in a manner analogous to the use of the serine primary hydroxyl.[35][36] However, due to the steric interference of the extra methyl group of threonine, the base member of the triad is the N-terminal amide which polarises an ordered water which, in turn, deprotonates the catalytic hydroxyl to increase its reactivity.[1][28] Similarly, there exist equivalent 'serine only' and 'cysteine only' configurations such as penicillin acylase G[i] and penicillin acylase V[j] which are evolutionarily related to the proteasome proteases. Again, these use their N-terminal amide as a base.[28]
Ser-cisSer-Lys
This unusual triad occurs only in one superfamily of amidases. Here, the lysine acts to polarise the middle serine.[45] The middle serine then forms two strong hydrogen bonds to the nucleophilic serine to activate it (one with the side chain hydroxyl and the other with the backbone amide). The middle serine is held in an unusual cis orientation to facilitate precise contacts with the other two triad residues. The triad is further unusual in that the lysine and cis-serine both act as the base in activating the catalytic serine, but the same lysine also performs the role of the acid member as well as making key structural contacts.[45][46]
Sec-His-Glu
The rare, but naturally occurring amino acid selenocysteine (Sec), can also be found as the nucleophile in some catalytic triads.[31] Selenocysteine is similar to cysteine, but contains a selenium atom instead of a sulfur. A selenocysteine residue is found in the active site of thioredoxin reductase, which uses the selenol group for reduction of disulfide in thioredoxin.[31]
Engineered triads
In addition to naturally occurring types of catalytic triads, protein engineering has been used to create enzyme variants with non-native amino acids, or entirely synthetic amino acids.[47] Catalytic triads have also been inserted into otherwise non-catalytic proteins, or protein mimics.[48]
Subtilisin (a serine protease) has had its oxygen nucleophile replaced with each of sulfur,[49][50] selenium,[51] or tellurium.[52] Cysteine and selenocysteine were inserted by mutagenesis, whereas the non-natural amino acid, tellurocysteine, was inserted using auxotrophic cells fed with synthetic tellurocysteine. These elements are all in the 16th periodic table column (chalcogens), so have similar properties.[53][54] In each case, changing the nucleophile lowered the enzyme's protease activity, but increased a new activity. A sulfur nucleophile improved the enzymes transferase activity (sometimes called subtiligase). Selenium and tellurium nucleophiles converted the enzyme into an oxidoreductase.[51][52] When the nucleophile of TEV protease was converted from cysteine to serine, its protease activity was strongly reduced, but it was possible to restore it by directed evolution.[55]
Non-catalytic proteins have been used as scaffolds, having catalytic triads inserted into them which were then improved by directed evolution. The Ser-His-Asp triad has been inserted into an antibody,[56] as well as a range of other proteins.[57] Similarly, catalytic triad mimics have been created in small organic molecules like diaryl diselenide,[58][59] and displayed on larger polymers like Merrifield resins,[60] and self-assembling short peptide nanostructures.[61]
Divergent evolution
Summarize
Perspective
The sophistication of the active site network causes residues involved in catalysis (and residues in contact with these) to be highly evolutionarily conserved.[62] However, many examples of divergent evolution in catalytic triads exist, both in the reaction catalysed, and the residues used in catalysis. The triad remains the core of the active site, but it is evolutionarily adapted to serve different functions.[63][64] Some proteins, called pseudoenzymes, have non-catalytic functions (e.g. regulation by inhibitory binding) and have accumulated mutations that inactivate their catalytic triad.[65]
Reaction changes
Catalytic triads perform covalent catalysis via an acyl-enzyme intermediate. If the intermediate is resolved by water, the result is hydrolysis of the substrate. However, if the intermediate is resolved by attack by a second substrate, then the enzyme acts as a transferase. For example, attack by an acyl group results in an acyltransferase reaction. Several families of transferase enzymes have evolved from hydrolases by adaptation to exclude water and favour attack of a second substrate.[66] In different members of the α/β-hydrolase superfamily, the Ser-His-Asp triad is tuned by surrounding residues to perform at least 17 different reactions.[39][67] Some of these reactions are also achieved with mechanisms that have altered formation, or resolution of the acyl-enzyme intermediate, or that don't proceed via an acyl-enzyme intermediate.[39]
Additionally, an alternative transferase mechanism has been evolved by amidophosphoribosyltransferase, which has two active sites.[k] In the first active site, a cysteine triad hydrolyses a glutamine substrate to release free ammonia. The ammonia then diffuses though an internal tunnel in the enzyme to the second active site, where it is transferred to a second substrate.[68][69]
Nucleophile changes

Divergent evolution of active site residues is slow, due to strong chemical constraints. Nevertheless, some protease superfamilies have evolved from one nucleophile to another. This can be inferred when a superfamily (with the same fold) contains families that use different nucleophiles.[55] Such nucleophile switches have occurred several times during evolutionary history, however the mechanisms by which this happen are still unclear.[17][55]
Within protease superfamilies that contain a mixture of nucleophiles (e.g. the PA clan), families are designated by their catalytic nucleophile (C=cysteine proteases, S=serine proteases).
| Superfamily | Families | Examples |
|---|---|---|
| PA clan | C3, C4, C24, C30, C37, C62, C74, C99 | TEV protease (Tobacco etch virus) |
| S1, S3, S6, S7, S29, S30, S31, S32, S39, S46, S55, S64, S65, S75 | Chymotrypsin (mammals, e.g. Bos taurus) | |
| PB clan | C44, C45, C59, C69, C89, C95 | Amidophosphoribosyltransferase precursor (Homo sapiens) |
| S45, S63 | Penicillin G acylase precursor (Escherichia coli) | |
| T1, T2, T3, T6 | Archaean proteasome, beta component (Thermoplasma acidophilum) | |
| PC clan | C26, C56 | Gamma-glutamyl hydrolase (Rattus norvegicus) |
| S51 | Dipeptidase E (Escherichia coli) | |
| PD clan | C46 | Hedgehog protein (Drosophila melanogaster) |
| N9, N10, N11 | Intein-containing V-type proton ATPase catalytic subunit A (Saccharomyces cerevisiae) | |
| PE clan | P1 | DmpA aminopeptidase (Brucella anthropi) |
| T5 | Ornithine acetyltransferase precursor (Saccharomyces cerevisiae) |
Pseudoenzymes
A further subclass of catalytic triad variants are pseudoenzymes, which have triad mutations that make them catalytically inactive, but able to function as binding or structural proteins.[71][72] For example, the heparin-binding protein Azurocidin is a member of the PA clan, but with a glycine in place of the nucleophile and a serine in place of the histidine.[73] Similarly, RHBDF1 is a homolog of the S54 family rhomboid proteases with an alanine in the place of the nucleophilic serine.[74][75] In some cases, pseudoenzymes may still have an intact catalytic triad but mutations in the rest of the protein remove catalytic activity. The CA clan contains catalytically inactive members with mutated triads (calpamodulin has lysine in place of its cysteine nucleophile) and with intact triads but inactivating mutations elsewhere (rat testin retains a Cys-His-Asn triad).[76]
| Superfamily | Families containing pseudoenzymes | Examples |
|---|---|---|
| CA clan | C1, C2, C19 | Calpamodulin |
| CD clan | C14 | CFLAR |
| SC clan | S9, S33 | Neuroligin |
| SK clan | S14 | ClpR |
| SR clan | S60 | Serotransferrin domain 2 |
| ST clan | S54 | RHBDF1 |
| PA clan | S1 | Azurocidin 1 |
| PB clan | T1 | PSMB3 |
Convergent evolution
Summarize
Perspective
Evolutionary convergence of serine and cysteine protease towards the same catalytic triads organisation of acid-base-nucleophile in different protease superfamilies. Shown are the triads of subtilisin,[l] prolyl oligopeptidase,[m] TEV protease,[a] and papain.[d] (PDB: 1ST2, 1LVM, 3EQ8, 1PE6)
The enzymology of proteases provides some of the clearest known examples of convergent evolution at a molecular level. The same geometric arrangement of triad residues occurs in over 20 separate enzyme superfamilies. Each of these superfamilies is the result of convergent evolution for the same triad arrangement within a different structural fold. This is because there are limited productive ways to arrange three triad residues, the enzyme backbone and the substrate. These examples reflect the intrinsic chemical and physical constraints on enzymes, leading evolution to repeatedly and independently converge on equivalent solutions.[1][2]
Cysteine and serine hydrolases
The same triad geometries been converged upon by serine proteases such as the chymotrypsin[c] and subtilisin superfamilies. Similar convergent evolution has occurred with cysteine proteases such as viral C3 protease and papain[d] superfamilies. These triads have converged to almost the same arrangement due to the mechanistic similarities in cysteine and serine proteolysis mechanisms.[2]
Families of cysteine proteases
| Superfamily | Families | Examples |
|---|---|---|
| CA | C1, C2, C6, C10, C12, C16, C19, C28, C31, C32, C33, C39, C47, C51, C54, C58, C64, C65, C66, C67, C70, C71, C76, C78, C83, C85, C86, C87, C93, C96, C98, C101 | Papain (Carica papaya) and calpain (Homo sapiens) |
| CD | C11, C13, C14, C25, C50, C80, C84 | Caspase-1 (Rattus norvegicus) and separase (Saccharomyces cerevisiae) |
| CE | C5, C48, C55, C57, C63, C79 | Adenain (human adenovirus type 2) |
| CF | C15 | Pyroglutamyl-peptidase I (Bacillus amyloliquefaciens) |
| CL | C60, C82 | Sortase A (Staphylococcus aureus) |
| CM | C18 | Hepatitis C virus peptidase 2 (hepatitis C virus) |
| CN | C9 | Sindbis virus-type nsP2 peptidase (sindbis virus) |
| CO | C40 | Dipeptidyl-peptidase VI (Lysinibacillus sphaericus) |
| CP | C97 | DeSI-1 peptidase (Mus musculus) |
| PA | C3, C4, C24, C30, C37, C62, C74, C99 | TEV protease (Tobacco etch virus) |
| PB | C44, C45, C59, C69, C89, C95 | Amidophosphoribosyltransferase precursor (Homo sapiens) |
| PC | C26, C56 | Gamma-glutamyl hydrolase (Rattus norvegicus) |
| PD | C46 | Hedgehog protein (Drosophila melanogaster) |
| PE | P1 | DmpA aminopeptidase (Brucella anthropi) |
| unassigned | C7, C8, C21, C23, C27, C36, C42, C53, C75 |
Families of serine proteases
| Superfamily | Families | Examples |
|---|---|---|
| SB | S8, S53 | Subtilisin (Bacillus licheniformis) |
| SC | S9, S10, S15, S28, S33, S37 | Prolyl oligopeptidase (Sus scrofa) |
| SE | S11, S12, S13 | D-Ala-D-Ala peptidase C (Escherichia coli) |
| SF | S24, S26 | Signal peptidase I (Escherichia coli) |
| SH | S21, S73, S77, S78, S80 | Cytomegalovirus assemblin (human herpesvirus 5) |
| SJ | S16, S50, S69 | Lon-A peptidase (Escherichia coli) |
| SK | S14, S41, S49 | Clp protease (Escherichia coli) |
| SO | S74 | Phage GA-1 neck appendage CIMCD self-cleaving protein (Bacillus phage GA-1) |
| SP | S59 | Nucleoporin 145 (Homo sapiens) |
| SR | S60 | Lactoferrin (Homo sapiens) |
| SS | S66 | Murein tetrapeptidase LD-carboxypeptidase (Pseudomonas aeruginosa) |
| ST | S54 | Rhomboid-1 (Drosophila melanogaster) |
| PA | S1, S3, S6, S7, S29, S30, S31, S32, S39, S46, S55, S64, S65, S75 | Chymotrypsin A (Bos taurus) |
| PB | S45, S63 | Penicillin G acylase precursor (Escherichia coli) |
| PC | S51 | Dipeptidase E (Escherichia coli) |
| PE | P1 | DmpA aminopeptidase (Brucella anthropi) |
| unassigned | S48, S62, S68, S71, S72, S79, S81 |
Threonine proteases
Threonine proteases use the amino acid threonine as their catalytic nucleophile. Unlike cysteine and serine, threonine is a secondary hydroxyl (i.e. has a methyl group). This methyl group greatly restricts the possible orientations of triad and substrate as the methyl clashes with either the enzyme backbone or histidine base.[2] When the nucleophile of a serine protease was mutated to threonine, the methyl occupied a mixture of positions, most of which prevented substrate binding.[77] Consequently, the catalytic residue of a threonine protease is located at its N-terminus.[2]
Two evolutionarily independent enzyme superfamilies with different protein folds are known to use the N-terminal residue as a nucleophile: Superfamily PB (proteasomes using the Ntn fold)[35] and Superfamily PE (acetyltransferases using the DOM fold)[36] This commonality of active site structure in completely different protein folds indicates that the active site evolved convergently in those superfamilies.[2][28]
Families of threonine proteases
| Superfamily | Families | Examples |
|---|---|---|
| PB clan | T1, T2, T3, T6 | Archaean proteasome, beta component (Thermoplasma acidophilum) |
| PE clan | T5 | Ornithine acetyltransferase (Saccharomyces cerevisiae) |
See also
References
Wikiwand - on
Seamless Wikipedia browsing. On steroids.