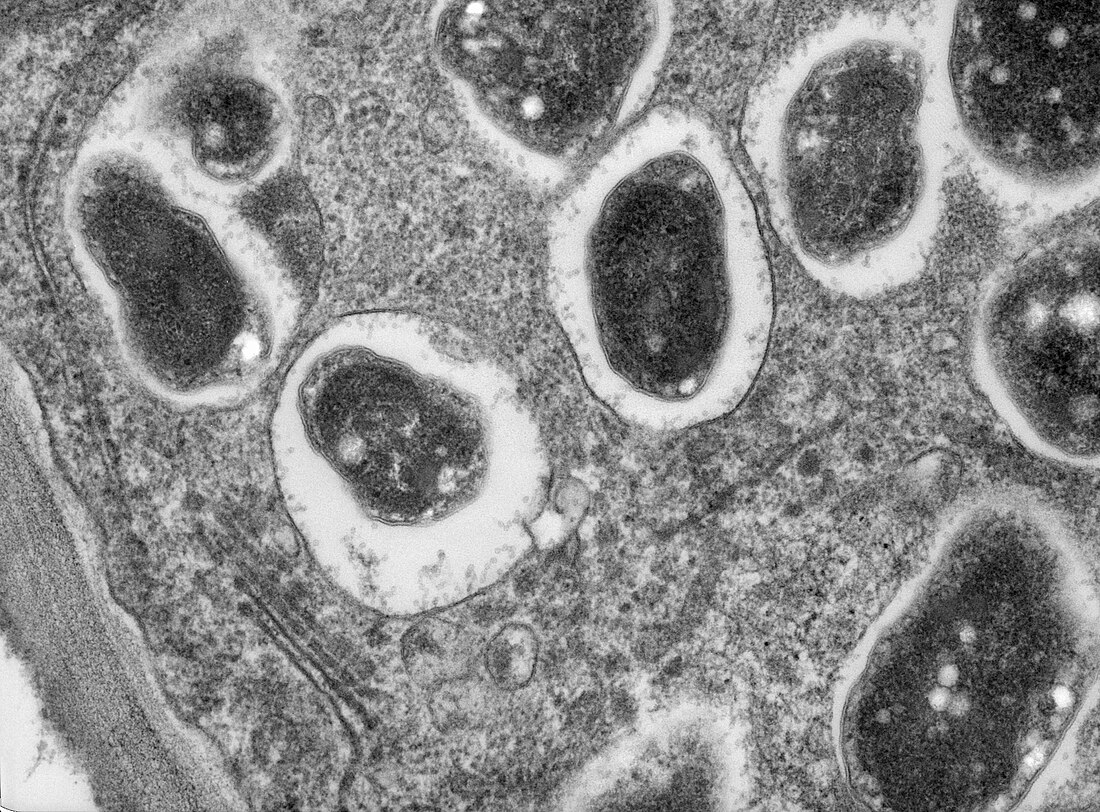
Bradyrhizobium japonicum
Species of bacterium From Wikipedia, the free encyclopedia
Bradyrhizobium japonicum is a species of legume-root nodulating, microsymbiotic nitrogen-fixing bacteria. The species is one of many Gram-negative, rod-shaped bacteria commonly referred to as rhizobia.[citation needed] Within that broad classification, which has three groups, taxonomy studies using DNA sequencing indicate that B. japonicum belongs within homology group II.[2]
| Bradyrhizobium japonicum | |
|---|---|
| Scientific classification | |
| Domain: | Bacteria |
| Kingdom: | Pseudomonadati |
| Phylum: | Pseudomonadota |
| Class: | Alphaproteobacteria |
| Order: | Hyphomicrobiales |
| Family: | Nitrobacteraceae |
| Genus: | Bradyrhizobium |
| Species: | B. japonicum |
| Binomial name | |
| Bradyrhizobium japonicum | |
| Synonyms | |
| |
Uses
Agriculture
B. japonicum is added to legume seed to improve crop yields,[3] particularly in areas where the bacterium is not native (e.g. Arkansas soils).[4] Often the inoculate is adhered to the seeds prior to planting using a sugar solution.[5]
Research
A strain of B. japonicum, USDA110, has been in use as a model organism since 1957.[citation needed] It is widely used to study molecular genetics, plant physiology, and plant ecology due to its relatively superior symbiotic nitrogen-fixation activity with soybean (i.e. compared to other rhizobia species). Its entire genome was sequenced in 2002, revealing that the species has a single circular chromosome with 9,105,828 base pairs.[6]
Metabolism
B. japonicum is able to degrade catechin with formation of phloroglucinol carboxylic acid, further decarboxylated to phloroglucinol, which is dehydroxylated to resorcinol and hydroxyquinol.[citation needed]
B. japonicum possess the nosRZDFYLX gene, which aides in denitrification and has two catalytic subunits - Cu-a and Cu-z (with several histidine residues). It manages an expression cascade that can sense oxygen gradients, termed 'FixJ-FixK2-FixK1.' FixJ positively regulates FixK2, which activates nitrogen respiration genes, as well as FixK1. FixK1 mutants are unable to respire from nitrogen due to a defective catatylic copper subunit (Cu-z) in nosRZDFYLX.[7]
Genetic transformation
Natural genetic transformation in bacteria is a sexual process involving transfer of DNA from one cell to another through the intervening medium, and the integration of the donor sequence into the recipient genome by homologous recombination. B. japonicum cells are able to undergo transformation.[8] They become competent for DNA uptake during late log phase.
References
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.