3C-like protease
Class of enzymes From Wikipedia, the free encyclopedia
The 3C-like protease (3CLpro) or main protease (Mpro), formally known as C30 endopeptidase or 3-chymotrypsin-like protease,[2] is the main protease found in coronaviruses. It cleaves the coronavirus polyprotein at eleven conserved sites. It is a cysteine protease and a member of the PA clan of proteases. It has a cysteine-histidine catalytic dyad at its active site and cleaves a Gln–(Ser/Ala/Gly) peptide bond.
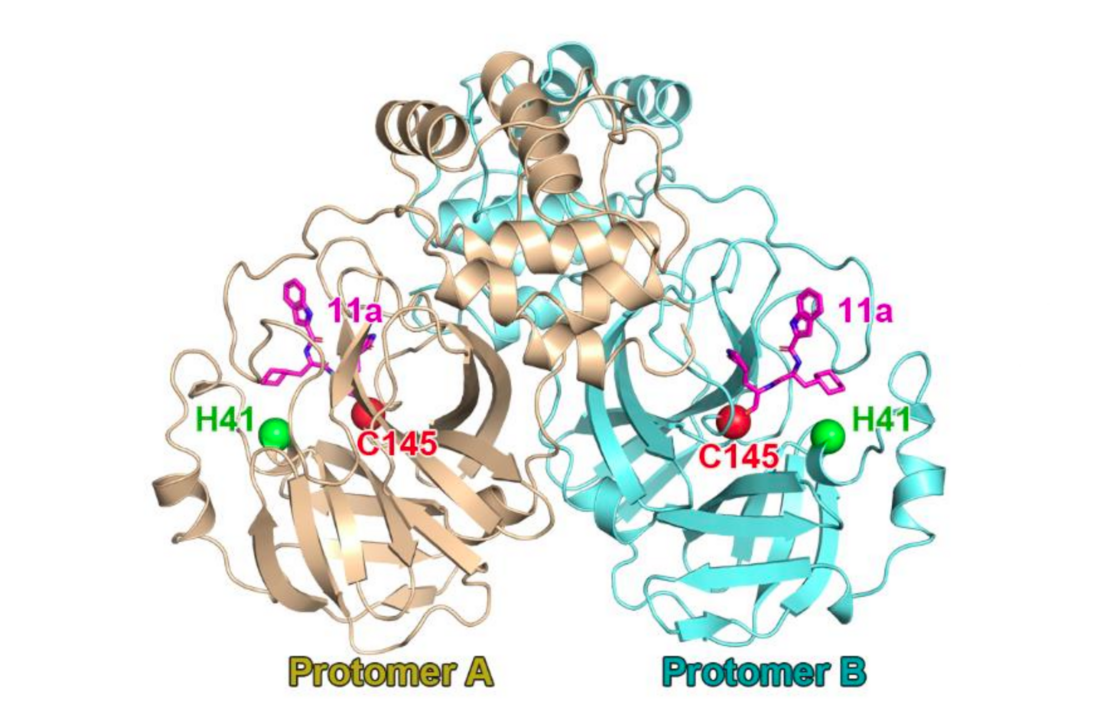
 SARS-CoV-2 main proteinase dimer with the catalytic dyad (H41; C145) in complex with a covalent peptidomimetic protease inhibitor ("11a", magenta). From PDB: 6LZE.[1] | |||||||||
| Identifiers | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| EC no. | 3.4.22.69 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
The Enzyme Commission refers to this family as SARS coronavirus main proteinase (Mpro; EC 3.4.22.69). The 3CL protease corresponds to coronavirus nonstructural protein 5 (nsp5). The "3C" in the common name refers to the 3C protease (3Cpro) which is a homologous protease found in picornaviruses.
Function
The 3C-like protease is able to catalytically cleave a peptide bond between a glutamine at position P1 and a small amino acid (serine, alanine, or glycine) at position P1'. The SARS coronavirus 3CLpro can for instance self-cleave the following peptides:[3][4][5]
TSAVLQ-SGFRK-NH2 and SGVTFQ-GKFKK are the two peptides corresponding to the two self-cleavage sites of the SARS 3C-like proteinase
The protease is important in the processing of the coronavirus replicase polyprotein (P0C6U8). It is the main protease in coronaviruses and corresponds to nonstructural protein 5 (nsp5).[6] It cleaves the coronavirus polyprotein at 11 conserved sites. The 3CL protease has a cysteine-histidine catalytic dyad at its active site.[4] The sulfur of the cysteine acts as a nucleophile and the imidazole ring of the histidine as a general base.[7]
| Position | Substrate preference |
|---|---|
| P5 | No strong preference |
| P4 | Small hydrophobic residues |
| P3 | Positively charged residue |
| P2 | High hydrophobicity and absence of beta-branch |
| P1 | Glutamine |
| P1' | Small residues |
| P2' | Small residues |
| P3' | No strong preference |
Nomenclature
Alternative names provided by the EC include 3CLpro, 3C-like protease, coronavirus 3C-like protease, Mpro, SARS 3C-like protease, SARS coronavirus 3CL protease, SARS coronavirus main peptidase, SARS coronavirus main protease, SARS-CoV 3CLpro enzyme, SARS-CoV main protease, SARS-CoV Mpro and severe acute respiratory syndrome coronavirus main protease.
As a treatment target
Summarize
Perspective


A number of protease inhibitors are being developed targeting 3CLpro and homologous 3Cpro, including CLpro-1, GC376, rupintrivir, lufotrelvir, PF-07321332, and AG7404.[11][12][13][14][1] The intravenous administered prodrug PF-07304814 (lufotrelvir) entered clinical trials in September 2020.[15]
After clinical trials, in December 2021, the oral medication nirmatrelvir (formerly PF-07321332) became commercially available under emergency use authorizations (EUA), as part of the nirmatrelvir/ritonavir combination therapy (brand name Paxlovid).[16][17] In May 2023, the medication got full FDA approval for high-risk adults, while children 12–18 were still covered under the EUA.[18]
The 3C-like protease inhibitor ensitrelvir received authorization to treat COVID-19 in Japan in 2022.[19][20]
In 2022, an ultralarge virtual screening campaign of 235 million molecules was able to identify a novel broad-spectrum inhibitor targeting the main protease of several coronaviruses. It is unusually not a peptidomimetic.[21]

Other 3C(-like) proteases
3C-like proteases (3C(L)pro) are widely found in (+)ssRNA viruses. All of them are cysteine proteases with a chymotrypsin-like fold (PA clan), using a catalytic dyad or triad. They share some general similarities on substrate specificity and inhibitor effectiveness. They are divided into subfamilies by sequence similarity, corresponding to the family of viruses they are found in:[22]
- This entry is the coronavirus 3CLpro.
- Picornaviridae have a picornavirus 3Cpro (EC 3.4.22.28; InterPro: IPR000199; MEROPS C03). This is the earliest-studied family. Examples include the ones found in poliovirus and in rhinovirus (both are members of genus Enterovirus).
- Caliciviridae have a 3CLpro (InterPro: IPR001665; MEROPS C37). Examples include the one found in Norwalk virus.
Additional members are known from Potyviridae and non-Coronaviridae Nidovirales.[23]
See also
- 3CLpro-1
- Carmofur
- COVID Moonshot
- Ebselen
- EDP-235
- Eganelisib
- GC376
- GRL-0617
- Iscartrelvir
- MK-7845
- Nirmatrelvir
- Olgotrelvir
- Leritrelvir (RAY1216)
- Rupintrivir
- SIM0417
- Theaflavin digallate
- Tollovir
- Y180
- Tetrahydrocannabutol
References
Further reading
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.