Transfer-messenger RNA
RNA family From Wikipedia, the free encyclopedia
Transfer-messenger RNA (abbreviated tmRNA, also known as 10Sa RNA and by its genetic name SsrA) is a bacterial RNA molecule with dual tRNA-like and messenger RNA-like properties. The tmRNA forms a ribonucleoprotein complex (tmRNP) together with Small Protein B (SmpB), Elongation Factor Tu (EF-Tu), and ribosomal protein S1. In trans-translation, tmRNA and its associated proteins bind to bacterial ribosomes which have stalled in the middle of protein biosynthesis, for example when reaching the end of a messenger RNA which has lost its stop codon. The tmRNA is remarkably versatile: it recycles the stalled ribosome, adds a proteolysis-inducing tag to the unfinished polypeptide, and facilitates the degradation of the aberrant messenger RNA.[1] In the majority of bacteria these functions are carried out by standard one-piece tmRNAs. In other bacterial species, a permuted ssrA gene produces a two-piece tmRNA in which two separate RNA chains are joined by base-pairing.

Discovery and early work
tmRNA was first designated 10Sa RNA in 1979, after a mixed "10S" electrophoretic fraction of Escherichia coli RNA was further resolved into tmRNA and the similarly sized RNase P RNA (10Sb).[2] The presence of pseudouridine in the mixed 10S RNA hinted that tmRNA has modified bases found also in tRNA. The similarity at the 3' end of tmRNA to the T stem-loop of tRNA was first recognized upon sequencing ssrA from Mycobacterium tuberculosis.[3] Subsequent sequence comparison revealed the full tRNA-like domain (TLD) formed by the 5' and 3' ends of tmRNA, including the acceptor stem with elements like those in alanine tRNA that promote its aminoacylation by alanine-tRNA ligase.[4] It also revealed differences from tRNA: the anticodon arm is missing in tmRNA, and the D arm region is a loop without base pairs.
Structure
Summarize
Perspective
Secondary structure of the standard one-piece tmRNAs

The complete E. coli tmRNA secondary structure was elucidated by comparative sequence analysis and structural probing.[5][6] Watson-Crick and G-U base pairs were identified by comparing the bacterial tmRNA sequences using automated computational methods in combination with manual alignment procedures.[7][8] The accompanying figure shows the base pairing pattern of this prototypical tmRNA, which is organized into 12 phylogenetically supported helices (also called pairings P1 to P12), some divided into helical segments.
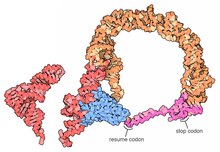
A prominent feature of every tmRNA is the conserved tRNA-like domain (TLD), composed of helices 1, 12, and 2a (analogs of the tRNA acceptor stem, T-stem and variable stem, respectively), and containing the 5' monophosphate and alanylatable 3' CCA ends. The mRNA-like region (MLR) is in standard tmRNA a large loop containing pseudoknots and a coding sequence (CDS) for the tag peptide, marked by the resume codon and the stop codon. The encoded tag peptide (ANDENYALAA in E. coli) varies among bacteria, perhaps depending on the set of proteases and adaptors available.[9]
tmRNAs typically contain four pseudoknots, one (pk1) upstream of the tag peptide CDS, and the other three pseudoknots (pk2 to pk4) downstream of the CDS. The pseudoknot regions, although generally conserved, are evolutionarily plastic. For example, in the (one-piece) tmRNAs of cyanobacteria, pk4 is substituted with two tandemly arranged smaller pseudoknots. This suggests that tmRNA folding outside the TLD can be important, yet the pseudoknot region lacks conserved residues and pseudoknots are among the first structures to be lost as ssrA sequences diverge in plastid and endosymbiont lineages. Base pairing in the three-pseudoknot region of E. coli tmRNA is disrupted during trans-translation.[7][10]
Two-piece tmRNAs
Circularly permuted ssrA has been reported in three major lineages: i) all alphaproteobacteria and the primitive mitochondria of jakobid protists, ii) two disjoint groups of cyanobacteria (Gloeobacter and a clade containing Prochlorococcus and many Synechococcus), and iii) some members of the betaproteobacteria (Cupriavidus and some Rhodocyclales).[11][12] All produce the same overall two-piece (acceptor and coding pieces) form, equivalent to the standard form nicked downstream of the reading frame. None retain more than two pseudoknots compared to the four (or more) of standard tmRNA.
Alphaproteobacteria have two signature sequences: replacement of the typical T-loop sequence TΨCRANY with GGCRGUA, and the sequence AACAGAA in the large loop of the 3´-terminal pseudoknot. In mitochondria, the MLR has been lost, and a remarkable re-permutation of mitochondrial ssrA results in a small one-piece product in Jakoba libera.[13]
The cyanobacteria provide the most plausible case for evolution of a permuted gene from a standard gene, due to remarkable sequence similarities between the two gene types as they occur in different Synechococcus strains.
tmRNA processing
Most tmRNAs are transcribed as larger precursors which are processed much like tRNA. Cleavage at the 5´ end is by ribonuclease P.[4] Multiple exonucleases can participate in the processing of the 3´ end of tmRNA, although RNase T and RNase PH are most effective.[14][15] Depending on the bacterial species, the 3'-CCA is either encoded or added by tRNA nucleotidyltransferase.
Similar processing at internal sites of permuted precursor tmRNA explains its physical splitting into two pieces. The two-piece tmRNAs have two additional ends whose processing must be considered. For alphaproteobacteria, one 5´ end is the unprocessed start site of transcription.[16] The far 3´ end may in some cases be the result of rho-independent termination.
Three-dimensional structures


High-resolution structures of the complete tmRNA molecules are currently unavailable and may be difficult to obtain due to the inherent flexibility of the MLR. In 2007, the crystal structure of the Thermus thermophilus TLD bound to the SmpB protein was obtained at 3 Å resolution. This structure shows that SmpB mimics the D stem and the anticodon of a canonical tRNA whereas helical section 2a of tmRNA corresponds to the variable arm of tRNA.[18] A cryo-electron microscopy study of tmRNA at an early stage of trans-translation shows the spatial relationship between the ribosome and the tmRNP (tmRNA bound to the EF-Tu protein). The TLD is located near the GTPase-associated center in the 50S ribosomal subunit; helix 5 and pseudoknots pk2 to pk4 form an arc around the beak of the 30S ribosomal subunit.[19]
Trans-translation
Summarize
Perspective

Coding by tmRNA was discovered in 1995[20] when Simpson and coworkers overexpressed the mouse cytokine IL-6 in E. coli and found multiple truncated cytokine-derived peptides each tagged at the carboxyl termini with the same 11-amino acid residue extension (A)ANDENYALAA. With the exception of the N-terminal alanine, which comes from the 3' end of tmRNA itself, this tag sequence was traced to a short open reading frame in E. coli tmRNA. Keiler, et al., recognized that the tag peptide confers proteolysis and proposed the trans-translation model for tmRNA action.[21]
While details of the trans-translation mechanism are under investigation it is generally agreed that tmRNA first occupies the empty A site of the stalled ribosome. Subsequently, the ribosome moves from the 3' end of the truncated messenger RNA onto the resume codon of the MLR, followed by a slippage-prone stage from where translation continues normally until the in-frame tmRNA stop codon is encountered. Trans-translation is essential in some bacterial species, whereas other bacteria require tmRNA to survive when subjected to stressful growth conditions.[22] It is believed that tmRNA can help the cell with antibiotic resistance by rescuing the ribosomes stalled by antibiotics.[23] Depending on the organism, the tag peptide may be recognized by a variety of proteases or protease adapters.[9]
Mobile genetic elements and the tmRNA gene
Summarize
Perspective

ssrA is both a target for some mobile DNAs and a passenger on others. It has been found interrupted by three types of mobile elements. By different strategies none of these disrupt gene function: group I introns remove themselves by self-splicing, rickettsial palindromic elements (RPEs) insert in innocuous sites, and integrase-encoding genomic islands split their target ssrA yet restore the split-off portion.[24][25][26][27]
Non-chromosomal ssrA was first detected in a genomic survey of mycobacteriophages (in 10% of the phages).[28] Other mobile elements including plasmids and genomic islands have been found bearing ssrA. One interesting case is Rhodobacter sphaeroides ATCC 17025, whose native tmRNA gene is disrupted by a genomic island; unlike all other genomic islands in tmRNA (or tRNA) genes this island has inactivated the native target gene without restoration, yet compensates by carrying its own tmRNA gene. A very unusual relative of ssrA is found in the lytic mycobacteriophage DS6A, that encodes little more than the TLD.
Mitochondrial tmRNAs (ssrA gene)
Summarize
Perspective
A mitochondrion-encoded, structurally reduced form of tmRNA (mt-tmRNA) was first postulated for the jakobid flagellate Reclinomonas americana.[11] Subsequently, the presence of a mitochondrial gene (ssrA) coding for tmRNA, as well as transcription and RNA processing sites were confirmed for all but one member of jakobids.[29][13] Functional evidence, i.e., mt-tmRNA Aminoacylation with alanine, is available for Jakoba libera.[13] More recently, ssrA was also identified in mitochondrial genomes of oomycetes.[30] Like in α-Proteobacteria (the ancestors of mitochondria), mt-tmRNAs are circularly permuted, two-piece RNA molecules, except in Jakoba libera where the gene has reverted to encoding a one-piece tmRNA conformation.[13]
Identification of ssrA in mitochondrial genomes

Mitochondrial tmRNA genes were initially recognized as short sequences that are conserved among jakobids and that have the potential to fold into a distinct tRNA-like secondary structure. With the availability of nine complete jakobid mtDNA sequences,[29] and a significantly improved covariance search tool (Infernal;[31][32][33]), a covariance model has been developed based on jakobid mitochondrial tmRNAs, which identified mitochondrial ssrA genes also in oomycete. At present, a total of 34 oomycete mt-tmRNAs have been detected across six genera: Albugo, Bremia, Phytophthora, Pseudoperonospora, Pythium and Saprolegnia. A covariance model built with both jakobid and oomycete sequences is now available at Rfam under the name ‘mt-tmRNA’.[30]
mt-tmRNA Structure
The standard bacterial tmRNA consists of a tRNA(Ala)-like domain (allowing addition of a non-encoded alanine to mRNAs that happen to lack a stop coding), and an mRNA-like domain coding for a protein tag that destines the polypeptide for proteolysis. The mRNA-like domain was lost in mt-tmRNAs. Comparative sequence analysis indicates features typical for mt-tmRNAs.[30] Most conserved is the primary sequence of the amino acyl acceptor stem. This portion of the molecule has an invariable A residue in the discriminator position and a G-U pair at position 3 (except in Seculamonas ecuadoriensis, which has a G-C pair); this position is the recognition site for alanyl tRNA synthase. P2 is a helix of variable length (3 to 10 base pairs) and corresponds to the anticodon stem of tRNAs, yet without an anticodon loop (as not required for tmRNA function). P2 stabilizes the tRNA-like structure, but four nucleotides invariant across oomycetes and jakobids suggest an additional, currently unidentified function. P3 has five base pairs and corresponds to the T-arm of tRNAs, yet with different consensus nucleotides both in the paired region and the loop. The T-loop sequence is conserved across oomycetes and jakobid, with only few deviations (e.g., Saprolegnia ferax). Finally, instead of the tRNA-like D-stem with a shortened three-nucleotide D-loop characteristic for bacterial tmRNAs, mitochondrial counterparts have a highly variable 5 to 14-nt long loop. The intervening sequence (Int.) of two-piece mt-tmRNAs is A+U rich and of irregular length (4-34 nt). ). For secondary structure models of one- and two-piece mt-tmRNAs see Figure 1.
mt-tmRNA processing and expression

RNA-Seq data of Phytophthora sojae show an expression level similar to that of neighboring mitochondrial tRNAs, and four major processing sites confirm the predicted termini of mature mt-tmRNA.[30] The tmRNA precursor molecule is likely processed by RNase P and a tRNA 3’ processing endonuclease (see Figure 2); the latter activity is assumed to lead to the removal of the intervening sequence. Following the addition of CCA at the 3’ discriminator nucleotide, the tmRNA can be charged by alanyl-tRNA synthetase with alanine.
See also
References
Further reading
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.