Top Qs
Timeline
Chat
Perspective
Rosetta@home
BOINC based volunteer computing project researching protein folding From Wikipedia, the free encyclopedia
Remove ads
Rosetta@home is a volunteer computing project researching protein structure prediction on the Berkeley Open Infrastructure for Network Computing (BOINC) platform, run by the Baker lab. Rosetta@home aims to predict protein–protein docking and design new proteins with the help of about fifty-five thousand active volunteered computers processing at over 487,946 GigaFLOPS on average as of September 19, 2020.[4] Foldit, a Rosetta@home videogame, aims to reach these goals with a crowdsourcing approach. Though much of the project is oriented toward basic research to improve the accuracy and robustness of proteomics methods, Rosetta@home also does applied research on malaria, Alzheimer's disease, and other pathologies.[5]
Remove ads
Like all BOINC projects, Rosetta@home uses idle computer processing resources from volunteers' computers to perform calculations on individual workunits. Completed results are sent to a central project server where they are validated and assimilated into project databases. The project is cross-platform, and runs on a wide variety of hardware configurations. Users can view the progress of their individual protein structure prediction on the Rosetta@home screensaver.
In addition to disease-related research, the Rosetta@home network serves as a testing framework for new methods in structural bioinformatics. Such methods are then used in other Rosetta-based applications, like RosettaDock or the Human Proteome Folding Project and the Microbiome Immunity Project, after being sufficiently developed and proven stable on Rosetta@home's large and diverse set of volunteer computers. Two especially important tests for the new methods developed in Rosetta@home are the Critical Assessment of Techniques for Protein Structure Prediction (CASP) and Critical Assessment of Prediction of Interactions (CAPRI) experiments, biennial experiments which evaluate the state of the art in protein structure prediction and protein–protein docking prediction, respectively. Rosetta consistently ranks among the foremost docking predictors, and is one of the best tertiary structure predictors available.[6]
With an influx of new users looking to participate in the fight against the COVID-19 pandemic, caused by SARS-CoV-2, Rosetta@home increased its computing power up to 1.7 PetaFlops as of March 28, 2020.[7][8] On September 9, 2020, Rosetta@home researchers published a paper describing 10 potent antiviral candidates against SARS-CoV-2. Rosetta@home contributed to this research and these antiviral candidates are heading towards Phase 1 clinical trials, which may begin in early 2022.[9][10][11][12] According to the Rosetta@home team, Rosetta volunteers contributed to the development of a nanoparticle vaccine.[9] This vaccine has been licensed and is known as the IVX-411 by Icosavax, which began a Phase I/II clinical trial in June 2021,[13] and GBP510 which is being developed by SK Bioscience and is already approved for a Phase III clinical trial in South Korea.[14][15]
NL-201, a cancer drug candidate that was first created at the Institute of Protein Design (IPD) and published in a January 2019 paper,[16] began a Phase 1 Human clinical trial in May 2021 with the support of Neoleukin Therapeutics, itself a spin-off from the IPD.[17] Rosetta@home played a role in the development of NL-201 and contributed with "forward folding" experiments that helped validate protein designs.[18]
Remove ads
Computing platform
Summarize
Perspective
The Rosetta@home application and the BOINC volunteer computing platform are available for the operating systems Windows, Linux, and macOS; BOINC also runs on several others, e.g., FreeBSD.[19] Participation in Rosetta@home requires a central processing unit (CPU) with a clock speed of at least 500 MHz, 200 megabytes of free disk space, 512 megabytes of physical memory, and Internet connectivity.[20] As of July 20, 2016, the current version of the Rosetta Mini application is 3.73.[21] The current recommended BOINC program version is 7.6.22.[19] Standard Hypertext Transfer Protocol (HTTP) (port 80) is used for communication between the user's BOINC client and the Rosetta@home servers at the University of Washington; HTTPS (port 443) is used during password exchange. Remote and local control of the BOINC client use port 31416 and port 1043, which might need to be specifically unblocked if they are behind a firewall.[22] Workunits containing data on individual proteins are distributed from servers located in the Baker lab at the University of Washington to volunteers' computers, which then calculate a structure prediction for the assigned protein. To avoid duplicate structure predictions on a given protein, each workunit is initialized with a random seed number. This gives each prediction a unique trajectory of descent along the protein's energy landscape.[23] Protein structure predictions from Rosetta@home are approximations of a global minimum in a given protein's energy landscape. That global minimum represents the most energetically favorable conformation of the protein, i.e., its native state.
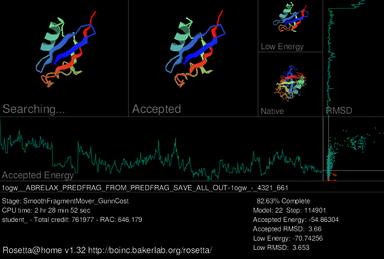
A primary feature of the Rosetta@home graphical user interface (GUI) is a screensaver which shows a current workunit's progress during the simulated protein folding process. In the upper-left of the current screensaver, the target protein is shown adopting different shapes (conformations) in its search for the lowest energy structure. Depicted immediately to the right is the structure of the most recently accepted. On the upper right the lowest energy conformation of the current decoy is shown; below that is the true, or native, structure of the protein if it has already been determined. Three graphs are included in the screensaver. Near the middle, a graph for the accepted model's thermodynamic free energy is displayed, which fluctuates as the accepted model changes. A graph of the accepted model's root-mean-square deviation (RMSD), which measures how structurally similar the accepted model is to the native model, is shown far right. On the right of the accepted energy graph and below the RMSD graph, the results from these two functions are used to produce an energy vs. RMSD plot as the model is progressively refined.[24]
Like all BOINC projects, Rosetta@home runs in the background of the user's computer, using idle computer power, either at or before logging into an account on the host operating system. The program frees resources from the CPU as they are needed by other applications so that normal computer use is unaffected. Many program settings can be specified via user account preferences, including: the maximum percentage of CPU resources the program can use (to control power consumption or heat production from a computer running at sustained capacity), the times of day during which the program can run, and many more.[citation needed]
Remove ads
Project significance
Summarize
Perspective
With the proliferation of genome sequencing projects, scientists can infer the amino acid sequence, or primary structure, of many proteins that carry out functions within the cell. To better understand a protein's function and aid in rational drug design, scientists need to know the protein's three-dimensional tertiary structure.

Protein 3D structures are currently determined experimentally via X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy. The process is slow (it can take weeks or even months to figure out how to crystallize a protein for the first time) and costly (around US$100,000 per protein).[25] Unfortunately, the rate at which new sequences are discovered far exceeds the rate of structure determination – out of more than 7,400,000 protein sequences available in the National Center for Biotechnology Information (NCBI) nonredundant (nr) protein database, fewer than 52,000 proteins' 3D structures have been solved and deposited in the Protein Data Bank, the main repository for structural information on proteins.[26] One of the main goals of Rosetta@home is to predict protein structures with the same accuracy as existing methods, but in a way that requires significantly less time and money. Rosetta@home also develops methods to determine the structure and docking of membrane proteins (e.g., G protein–coupled receptors (GPCRs)),[27] which are exceptionally difficult to analyze with traditional techniques like X-ray crystallography and NMR spectroscopy, yet represent the majority of targets for modern drugs.[28]
Progress in protein structure prediction is evaluated in the biannual Critical Assessment of Techniques for Protein Structure Prediction (CASP) experiment, in which researchers from around the world attempt to derive a protein's structure from the protein's amino acid sequence. High scoring groups in this sometimes competitive experiment are considered the de facto standard-bearers for what is the state of the art in protein structure prediction. Rosetta, the program on which Rosetta@home is based, has been used since CASP5 in 2002. In the 2004 CASP6 experiment, Rosetta made history by being the first to produce a close to atomic-level resolution, ab initio protein structure prediction in its submitted model for CASP target T0281.[29] Ab initio modeling is considered an especially difficult category of protein structure prediction, as it does not use information from structural homology and must rely on information from sequence homology and modeling physical interactions within the protein. Rosetta@home has been used in CASP since 2006, where it was among the top predictors in every category of structure prediction in CASP7.[30][31][32] These high quality predictions were enabled by the computing power made available by Rosetta@home volunteers.[33] Increasing computing power allows Rosetta@home to sample more regions of conformation space (the possible shapes a protein can assume), which, according to Levinthal's paradox, is predicted to increase exponentially with protein length.[citation needed]
Rosetta is also used in protein–protein docking prediction, which determines the structure of multiple complexed proteins, or quaternary structure. This type of protein interaction affects many cellular functions, including antigen–antibody and enzyme–inhibitor binding and cellular import and export. Determining these interactions is critical for drug design. Rosetta is used in the Critical Assessment of Prediction of Interactions (CAPRI) experiment, which evaluates the state of the protein docking field similar to how CASP gauges progress in protein structure prediction. The computing power made available by Rosetta@home's project volunteers has been cited as a major factor in Rosetta's performance in CAPRI 2007, where its docking predictions have been among the most accurate and complete.[34]
In early 2008, Rosetta was used to computationally design a protein with a function never before observed in nature.[35] This was inspired in part by the retraction of a high-profile paper from 2004 which originally described the computational design of a protein with improved enzymatic activity relative to its natural form.[36] The 2008 research paper from David Baker's group describing how the protein was made, which cited Rosetta@home for the computing resources it made available, represented an important proof of concept for this protein design method.[35] This type of protein design could have future applications in drug discovery, green chemistry, and bioremediation.[35]
The Rosetta computer program was cited in the 2024 Scientific Background to the Nobel Prize in Chemistry.[37]
Remove ads
Disease-related research
Summarize
Perspective
In addition to basic research in predicting protein structure, docking and design, Rosetta@home is also used in immediate disease-related research.[38] Numerous minor research projects are described in David Baker's Rosetta@home journal.[39] As of February 2014, information on recent publications and a short description of the work are being updated on the forum.[40] The forum thread is no longer used since 2016, and news on the research can be found on the general news section of the project.[41]
Alzheimer's disease
A component of the Rosetta software suite, RosettaDesign, was used to accurately predict which regions of amyloidogenic proteins were most likely to make amyloid-like fibrils.[42] By taking hexapeptides (six amino acid-long fragments) of a protein of interest and selecting the lowest energy match to a structure similar to that of a known fibril forming hexapeptide, RosettaDesign was able to identify peptides twice as likely to form fibrils as are random proteins.[43] Rosetta@home was used in the same study to predict structures for amyloid beta, a fibril-forming protein that has been postulated to cause Alzheimer's disease.[44] Preliminary but as yet unpublished results have been produced on Rosetta-designed proteins that may prevent fibrils from forming, although it is unknown whether it can prevent the disease.[45]
Anthrax
Another component of Rosetta, RosettaDock,[46][47][48] was used in conjunction with experimental methods to model interactions between three proteins—lethal factor (LF), edema factor (EF) and protective antigen (PA)—that make up anthrax toxin. The computer model accurately predicted docking between LF and PA, helping to establish which domains of the respective proteins are involved in the LF–PA complex. This insight was eventually used in research resulting in improved anthrax vaccines.[49][50]
Herpes simplex virus 1
RosettaDock was used to model docking between an antibody (immunoglobulin G) and a surface protein expressed by the cold sore virus, herpes simplex virus 1 (HSV-1) which serves to degrade the antiviral antibody. The protein complex predicted by RosettaDock closely agreed with the especially difficult-to-obtain experimental models, leading researchers to conclude that the docking method has potential to address some of the problems that X-ray crystallography has with modelling protein–protein interfaces.[51]
HIV
As part of research funded by a $19.4 million grant by the Bill & Melinda Gates Foundation,[52] Rosetta@home has been used in designing multiple possible vaccines for human immunodeficiency virus (HIV).[53][54]
Malaria
In research involved with the Grand Challenges in Global Health initiative,[55] Rosetta has been used to computationally design novel homing endonuclease proteins, which could eradicate Anopheles gambiae or otherwise render the mosquito unable to transmit malaria.[56] Being able to model and alter protein–DNA interactions specifically, like those of homing endonucleases, gives computational protein design methods like Rosetta an important role in gene therapy (which includes possible cancer treatments).[38][57]
COVID-19
In 2020, the Rosetta molecular modelling suite was used to accurately predict the atomic-scale structure of the SARS-CoV-2 spike protein weeks before it could be measured in the lab.[58] On June 26 of 2020, the project announced it had succeeded in creating antiviral proteins that neutralize SARS-CoV-2 virions in the lab and that these experimental antiviral drugs are being optimized for animal testing trials.[59]
In a follow-up, a paper describing 10 SARS-CoV-2 miniprotein inhibitors was published in Science on September 9. Two of these inhibitors, LCB1 and LCB3, are several times more potent than the best monoclonal antibodies being developed against SARS-CoV-2, both on a molar and mass basis. In addition, the research suggests that these inhibitors retain their activity at elevated temperatures, are 20-fold smaller than an antibody and thus, have 20-fold more potential neutralizing sites, increasing the potential efficacy of a locally administered drug. The small size and high stability of the inhibitors is expected to make them adequate to a gel formulation that can be nasally applied or as a powder to be administered directly onto the respiratory system. The researchers will work on developing these inhibitors into therapeutics and prophylactics in the months ahead.[10] As of July 2021, these antiviral candidates were forecasted to begin clinical trials in early 2022 and had received funding from the Bill & Melinda Gates Foundation for preclinical and early clinical trials.[12] In animal testing trials, these antiviral candidates were effective against variants of concern including Alpha, Beta and Gamma.[12][60][61]
Rosetta@home was used to help screen the over 2 million SARS-CoV-2 Spike-binding proteins that were computationally designed, and thus, contributed to this research.[62][63]
Per the Rosetta@home team at the Institute of Protein Design, Rosetta@home volunteers contributed to the development of antiviral drug candidates[10] and to a protein nanoparticle vaccine.[64] The IVX-411 vaccine is already on a Phase 1 clinical trial run by Icosavax[13] while the same vaccine, licensed to another manufacturer and under the name GBP510, has been approved in South Korea for a Phase III trial run by SK Bioscience.[15][14] The candidate antivirals are also going towards Phase 1 clinical trials.[9]
Cancer
Rosetta@home researchers have designed an IL-2 receptor agonist called Neoleukin-2/15 that does not interact with the alpha subunit of the receptor. Such immunity signal molecules are useful in cancer treatment. While the natural IL-2 suffers from toxicity due to an interaction with the alpha subunit, the designed protein is much safer, at least in animal models.[16] Rosetta@home contributed in "forward folding experiments" which helped validate designs.[18]
In a September 2020 feature in the New Yorker, David Baker stated that Neoleukin-2/15 would begin human clinical trials "later this year". Neoleukin-2/15 is being developed by Neoleukin, a spin-off company from the Baker lab.[65] In December 2020, Neoleukin announced it would be submitting an Investigational New Drug application with the Food and Drug Administration in order to begin a Phase 1 clinical trial of NL-201, which is a further development of Neoleukin-2/15. A similar application was submitted in Australia and Neoleukin hopes to enrol up 120 participants on the Phase 1 clinical trial.[66] The Phase 1 human clinical trial began on May 5, 2021.[17]
Remove ads
Rosetta software
Summarize
Perspective
Rosetta is the software responsible for performing structure prediction in Rosetta@home. Besides a BOINC cluster, Rosetta can run on a single local computer, or on a local supercomputer. Similar to other bioinformatic programs, there are online public servers offering to run Rosetta from a web interface.[71] The software is freely licensed to the academic community and available to pharmaceutical companies for a fee.[72]
Originally introduced by the Baker laboratory at the University of Washington in 1998 as an ab initio approach to structure prediction, Rosetta has since branched into several development streams and distinct services, providing features such as macromolecular docking and protein design.[67] Many of the graduate students and other researchers involved in Rosetta's initial development have since moved to other universities and research institutions, and subsequently enhanced different parts of the Rosetta project.
The Rosetta platform derives its name from the Rosetta Stone, as it attempts to decipher the structural "meaning" of proteins' amino acid sequences.[73] Development of the Rosetta code is done by Rosetta Commons.[72] Rosetta participates in CASP and CAPRI.
Rosetta was rewritten in C++ to allow easier development than that allowed by its original version, which was written in Fortran. This new version is object-oriented, and was released to Rosetta@Home February 8, 2008.[21][74]
RosettaDesign

RosettaDesign, a computing approach to protein design based on Rosetta, began in 2000 with a study in redesigning the folding pathway of Protein G.[75] In 2002 RosettaDesign was used to design Top7, a 93-amino acid long α/β protein that had an overall fold never before recorded in nature. This new conformation was predicted by Rosetta to within 1.2 Å RMSD of the structure determined by X-ray crystallography, representing an unusually accurate structure prediction.[76] Rosetta and RosettaDesign earned widespread recognition by being the first to design and accurately predict the structure of a novel protein of such length, as reflected by the 2002 paper describing the dual approach prompting two positive letters in the journal Science,[77][78] and being cited by more than 240 other scientific articles.[79] The visible product of that research, Top7, was featured as the RCSB PDB's 'Molecule of the Month' in October 2006;[80] a superposition of the respective cores (residues 60–79) of its predicted and X-ray crystal structures are featured in the Rosetta@home logo.[29]
Brian Kuhlman, a former postdoctoral associate in David Baker's lab and now an associate professor at the University of North Carolina, Chapel Hill,[81] offers RosettaDesign as an online service.[82]
RosettaDock
RosettaDock was added to the Rosetta software suite during the first CAPRI experiment in 2002 as the Baker laboratory's algorithm for protein–protein docking prediction.[83] In that experiment, RosettaDock made a high-accuracy prediction for the docking between streptococcal pyogenic exotoxin A and a T cell-receptor β-chain, and a medium accuracy prediction for a complex between porcine α-amylase and a camelid antibody. While the RosettaDock method only made two acceptably accurate predictions out of seven possible, this was enough to rank it seventh out of nineteen prediction methods in the first CAPRI assessment.[83]
Development of RosettaDock diverged into two branches for subsequent CAPRI rounds as Jeffrey Gray, who laid the groundwork for RosettaDock while at the University of Washington, continued working on the method in his new position at Johns Hopkins University. Members of the Baker laboratory further developed RosettaDock in Gray's absence. The two versions differed slightly in side-chain modeling, decoy selection and other areas.[48][84] Despite these differences, both the Baker and Gray methods performed well in the second CAPRI assessment, placing fifth and seventh respectively out of 30 predictor groups.[85] Jeffrey Gray's RosettaDock server is available as a free docking prediction service for non-commercial use.[86]
In October 2006, RosettaDock was integrated into Rosetta@home. The method used a fast, crude docking model phase using only the protein backbone. This was followed by a slow full-atom refinement phase in which the orientation of the two interacting proteins relative to each other, and side-chain interactions at the protein–protein interface, were simultaneously optimized to find the lowest energy conformation.[87] The vastly increased computing power afforded by the Rosetta@home network, combined with revised fold-tree representations for backbone flexibility and loop modeling, made RosettaDock sixth out of 63 prediction groups in the third CAPRI assessment.[6][34]
Robetta
The Robetta (Rosetta Beta) server is an automated protein structure prediction service offered by the Baker laboratory for non-commercial ab initio and comparative modeling.[88] It has participated as an automated prediction server in the biannual CASP experiments since CASP5 in 2002, performing among the best in the automated server prediction category.[89] Robetta has since competed in CASP6 and 7, where it did better than average among both automated server and human predictor groups.[32][90][91] It also participates in the CAMEO3D continuous evaluation. Robetta tasks run on Baker lab servers, Janelia Research Campus machines, and Rosetta@home participant computers.[88]
In modeling protein structure as of CASP6, Robetta first searches for structural homologs using BLAST, PSI-BLAST, and 3D-Jury, then parses the target sequence into its individual domains, or independently folding units of proteins, by matching the sequence to structural families in the Pfam database. Domains with structural homologs then follow a "template-based model" (i.e., homology modeling) protocol. Here, the Baker laboratory's in-house alignment program, K*sync, produces a group of sequence homologs, and each of these is modeled by the Rosetta de novo method to produce a decoy (possible structure). The final structure prediction is selected by taking the lowest energy model as determined by a low-resolution Rosetta energy function. For domains that have no detected structural homologs, a de novo protocol is followed in which the lowest energy model from a set of generated decoys is selected as the final prediction. These domain predictions are then connected together to investigate inter-domain, tertiary-level interactions within the protein. Finally, side-chain contributions are modeled using a protocol for Monte Carlo conformational search.[92]
In CASP8, Robetta was augmented to use Rosetta's high resolution all-atom refinement method,[93] the absence of which was cited as the main cause for Robetta being less accurate than the Rosetta@home network in CASP7.[33] In CASP11, a way to predict the protein contact map by co-evolution of residues in related proteins called GREMLIN was added, allowing for more de novo fold successes.[94]
Other Rosetta servers
Rosetta is available as an online service from a number of other public servers. ROSIE offers a variety of functions from RNA structure prediction and design to ligand docking and antibody modeling.[95]
Foldit
On May 9, 2008, after Rosetta@home users suggested an interactive version of the volunteer computing program, the Baker lab publicly released Foldit, an online protein structure prediction game based on the Rosetta platform.[96] As of September 25, 2008[update], Foldit had over 59,000 registered users.[97] The game gives users a set of controls (for example, shake, wiggle, rebuild) to manipulate the backbone and amino acid side chains of the target protein into more energetically favorable conformations. Users can work on solutions individually as soloists or collectively as evolvers, accruing points under either category as they improve their structure predictions.[98]
Foldit can work as a GUI frontend to Rosetta under a tailored "professional mode".[71]
RoseTTAFold
RoseTTAFold, which is inspired by AlphaFold, uses a neural network to predict the distance and orientation between residues. These predictions guide Rosetta software in producing a structure. RoseTTAFold is open source under the MIT license.[99]
Non-Baker lab branches
The Jianyi Yang lab in China offers a modified version of Rosetta termed tr-RosettaX2 (transform-restrained Rosetta).[100] It uses a deep learning-based contact prediction method different from RoseTTAFold to guide the usual Rosetta folding algorithm. trRosetta predates RoseTTAFold.[101]
Remove ads
Comparison to similar volunteer computing projects
Summarize
Perspective
There are several volunteer computed projects which have study areas similar to those of Rosetta@home, but differ in their research approach:
Folding@home
Of all the major volunteer computing projects involved in protein research, Folding@home is the only one not using the BOINC platform.[102][103][104] Both Rosetta@home and Folding@home study protein misfolding diseases such as Alzheimer's disease, but Folding@home does so much more exclusively.[105][106] Folding@home almost exclusively uses all-atom molecular dynamics models to understand how and why proteins fold (or potentially misfold, and subsequently aggregate to cause diseases).[107][108] In other words, Folding@home's strength is modeling the process of protein folding, while Rosetta@home's strength is computing protein design and predicting protein structure and docking.
Some of Rosetta@home's results are used as the basis for some Folding@home projects. Rosetta provides the most likely structure, but it is not definite if that is the form the molecule takes or whether or not it is viable. Folding@home can then be used to verify Rosetta@home's results, and can provide added atomic-level information, and details of how the molecule changes shape.[108][109]
The two projects also differ significantly in their computing power and host diversity. Averaging about 6,650 teraFLOPS from a host base of central processing units (CPUs), graphics processing units (GPUs), and (formerly) PS3s,[110] Folding@home has nearly 108 times more computing power than Rosetta@home.[111]
World Community Grid
Both Phase I and Phase II of the Human Proteome Folding Project (HPF), a subproject of World Community Grid, have used the Rosetta program to make structural and functional annotations of various genomes.[112][113] Although he now uses it to create databases for biologists, Richard Bonneau, head scientist of the Human Proteome Folding Project, was active in the original development of Rosetta at David Baker's laboratory while obtaining his PhD.[114] More information on the relationship between the HPF1, HPF2 and Rosetta@home can be found on Richard Bonneau's website.[115]
Predictor@home
Like Rosetta@home, Predictor@home specialized in protein structure prediction.[116] While Rosetta@home uses the Rosetta program for its structure prediction, Predictor@home used the dTASSER methodology.[117] In 2009, Predictor@home shut down.
Other protein related volunteer computing projects on BOINC include QMC@home, Docking@home, POEM@home, SIMAP, and TANPAKU. RALPH@home, the Rosetta@home alpha project which tests new application versions, work units, and updates before they move on to Rosetta@home, runs on BOINC also.[118]
Remove ads
Volunteer contributions
Summarize
Perspective
Rosetta@home depends on computing power donated by individual project members for its research. As of March 28, 2020[update], about 53,000 users from 150 countries were active members of Rosetta@home, together contributing idle processor time from about 54,800 computers for a combined average performance of over 1.7 PetaFLOPS.[111][119]

Users are granted BOINC credits as a measure of their contribution. The credit granted for each workunit is the number of decoys produced for that workunit multiplied by the average claimed credit for the decoys submitted by all computer hosts for that workunit. This custom system was designed to address significant differences between credit granted to users with the standard BOINC client and an optimized BOINC client, and credit differences between users running Rosetta@home on Windows and Linux operating systems.[120] The amount of credit granted per second of CPU work is lower for Rosetta@home than most other BOINC projects.[121] Rosetta@home is thirteenth out of over 40 BOINC projects in terms of total credit.[122]
Rosetta@home users who predict protein structures submitted for the CASP experiment are acknowledged in scientific publications regarding their results.[33] Users who predict the lowest energy structure for a given workunit are featured on the Rosetta@home homepage as Predictor of the Day, along with any team of which they are a member.[123] A User of the Day is chosen randomly each day to be on the homepage also, from among users who have made a Rosetta@home profile.[124]
Remove ads
References
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
Remove ads


