MARK4
Protein-coding gene in the species Homo sapiens From Wikipedia, the free encyclopedia
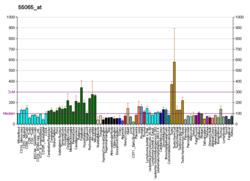
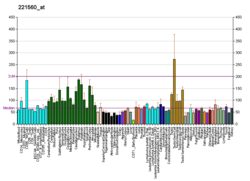
MAP/microtubule affinity-regulating kinase 4 is an enzyme that in humans is encoded by the MARK4 gene.[5][6][7] MARK4 belongs to the family of serine/threonine kinases that phosphorylate microtubule-associated proteins (MAP) causing their detachment from microtubules.[8] Detachment thereby increases microtubule dynamics and facilitates a number of cell activities including cell division, cell cycle control, cell polarity determination, and cell shape alterations.[9]
There are four members of the MARK protein family, MARK1-4, which are highly conserved. MARK4 kinase has been shown to be involved in microtubule organization in neuronal cells. Levels of MARK4 are elevated in Alzheimer's disease and may contribute to the pathological phosphorylation of tau protein in this disease.
Interactions
MARK4 has been shown to interact with USP9X[10] and Ubiquitin C.[10]
References
Further reading
Wikiwand - on
Seamless Wikipedia browsing. On steroids.