DNA clamp
Protein family From Wikipedia, the free encyclopedia
A DNA clamp, also known as a sliding clamp, is a protein complex that serves as a processivity-promoting factor in DNA replication. As a critical component of the DNA polymerase III holoenzyme, the clamp protein binds DNA polymerase and prevents this enzyme from dissociating from the template DNA strand. The clamp-polymerase protein–protein interactions are stronger and more specific than the direct interactions between the polymerase and the template DNA strand; because one of the rate-limiting steps in the DNA synthesis reaction is the association of the polymerase with the DNA template, the presence of the sliding clamp dramatically increases the number of nucleotides that the polymerase can add to the growing strand per association event. The presence of the DNA clamp can increase the rate of DNA synthesis up to 1,000-fold compared with a nonprocessive polymerase.[2]
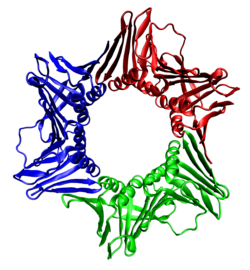
Top and side views of a homotrimer of the human PCNA sliding clamp (rainbow colored, N-terminus = blue, C-terminus = red) with double stranded DNA modeled through the central pore (magenta).[1]


Structure
Summarize
Perspective
The DNA clamp is an α+β protein that assembles into a multimeric, six-domain ring structure that completely encircles the DNA double helix as the polymerase adds nucleotides to the growing strand.[3] Each domain is in turn made of two β-α-β-β-β structural repeats.[4] The DNA clamp assembles on the DNA at the replication fork and "slides" along the DNA with the advancing polymerase, aided by a layer of water molecules in the central pore of the clamp between the DNA and the protein surface. Because of the toroidal shape of the assembled multimer, the clamp cannot dissociate from the template strand without also dissociating into monomers.
The DNA clamp fold is found in bacteria, archaea, eukaryotes and some viruses. In bacteria, the sliding clamp is a homodimer composed of two identical beta subunits of DNA polymerase III and hence is referred to as the beta clamp. In archaea[5] and eukaryotes, it is a trimer composed of three molecules of PCNA. The T4 bacteriophage also uses a sliding clamp, called gp45 that is a trimer similar in structure to PCNA but lacks sequence homology to either PCNA or the bacterial beta clamp.[3]
| Taxon | Sliding clamp protein | Multimer state | Associated polymerase |
|---|---|---|---|
| Bacteria | beta subunit of pol III | dimer | DNA polymerase III |
| Archaea | archaeal PCNA | trimer | PolD |
| Eukaryote | PCNA | trimer | DNA polymerase delta |
| Caudovirales | IPR004190 | trimer | Phage polymerase (e.g. T4) |
| Herpesviridae | non-clamp processivity factor | monomer | Virus-encoded polymerase |
Bacterial
| DNA polymerase III subunit beta | |||||||
|---|---|---|---|---|---|---|---|
 | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | dnaN | ||||||
| Entrez | 948218 | ||||||
| PDB | 1MMI | ||||||
| RefSeq (Prot) | NP_418156 | ||||||
| UniProt | P0A988 | ||||||
| Other data | |||||||
| EC number | 2.7.7.7 | ||||||
| Chromosome | MG1655: 3.88 - 3.88 Mb | ||||||
| |||||||
The beta clamp is a specific DNA clamp and a subunit of the DNA polymerase III holoenzyme found in bacteria. Two beta subunits are assembled around the DNA by the gamma subunit and ATP hydrolysis; this assembly is called the pre-initiation complex. After assembly around the DNA, the beta subunits' affinity for the gamma subunit is replaced by an affinity for the alpha and epsilon subunits, which together create the complete holoenzyme.[7][8][9] DNA polymerase III is the primary enzyme complex involved in prokaryotic DNA replication.
The gamma complex of DNA polymerase III, composed of γδδ'χψ subunits, catalyzes ATP to chaperone two beta subunits to bind to DNA. Once bound to DNA, the beta subunits can freely slide along double stranded DNA. The beta subunits in turn bind the αε polymerase complex. The α subunit possesses DNA polymerase activity and the ε subunit is a 3’-5’ exonuclease.[9]
The beta chain of bacterial DNA polymerase III is composed of three topologically equivalent domains (N-terminal, central, and C-terminal). Two beta chain molecules are tightly associated to form a closed ring encircling duplex DNA.
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
As a drug target
Certain NSAIDs (carprofen, bromfenac, and vedaprofen) exhibit some suppression of bacterial DNA replication by inhibiting bacterial DNA clamp.[10]
Eukaryotic and archaeal
The sliding clamp in eukaryotes is assembled from a specific subunit of DNA polymerase delta called the proliferating cell nuclear antigen (PCNA). The N-terminal and C-terminal domains of PCNA are topologically identical. Three PCNA molecules are tightly associated to form a closed ring encircling duplex DNA.
The sequence of PCNA is well conserved between plants, animals and fungi, indicating a strong selective pressure for structure conservation, and suggesting that this type of DNA replication mechanism is conserved throughout eukaryotes.[12][13] In eukaryotes, a homologous, heterotrimeric "9-1-1 clamp" made up of RAD9-RAD1-HUS1 (911) is responsible for DNA damage checkpoint control.[14] This 9-1-1 clamp mounts onto DNA in the opposite direction.[15]
Archaea, probable evolutionary precursor of eukaryotes, also universally have at least one PCNA gene. This PCNA ring works with PolD, the single eukaryotic-like DNA polymerase in archaea responsible for multiple functions from replication to repair. Some unusual species have two or even three PCNA genes, forming heterotrimers or distinct specialized homotrimers.[16] Archaeons also share with eukaryotes the PIP (PCNA-interacting protein) motif, but a wider variety of such proteins performing different functions are found.[17]
PCNA is also appropriated by some viruses. The giant virus genus Chlorovirus, with PBCV-1 as a representative, carries in its genome two PCNA genes (Q84513, O41056) and a eukaryotic-type DNA polymerase.[18] Members of Baculoviridae also encode a PCNA homolog (P11038).[19]
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
Caudoviral
| DNA polymerase accessory protein 45 | |||||||
|---|---|---|---|---|---|---|---|
 | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | gp45 | ||||||
| Entrez | 1258821 | ||||||
| PDB | 1CZD | ||||||
| RefSeq (Prot) | NP_049666 | ||||||
| UniProt | P04525 | ||||||
| Other data | |||||||
| EC number | 2.7.7.7 | ||||||
| Chromosome | 1: 0.03 - 0.03 Mb | ||||||
| |||||||
The viral gp45 sliding clamp subunit protein contains two domains. Each domain consists of two alpha helices and two beta sheets – the fold is duplicated and has internal pseudo two-fold symmetry.[21] Three gp45 molecules are tightly associated to form a closed ring encircling duplex DNA.
|
| ||||||||||||||||||||||||||||||||||||||||||||
Herpesviral
Some members of Herpesviridae encode a protein that has a DNA clamp fold but does not associate into a ring clamp. The two-domain protein does, however, associate with the viral DNA polymerase and also acts to increase processivity.[22] As it does not form a ring, it does not need a clamp loader to be attached to DNA.[23]
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Assembly
Sliding clamps are loaded onto their associated DNA template strands by specialized proteins known as "sliding clamp loaders", which also disassemble the clamps after replication has completed. The binding sites for these initiator proteins overlap with the binding sites for the DNA polymerase, so the clamp cannot simultaneously associate with a clamp loader and with a polymerase. Thus the clamp will not be actively disassembled while the polymerase remains bound. DNA clamps also associate with other factors involved in DNA and genome homeostasis, such as nucleosome assembly factors, Okazaki fragment ligases, and DNA repair proteins. All of these proteins also share a binding site on the DNA clamp that overlaps with the clamp loader site, ensuring that the clamp will not be removed while any enzyme is still working on the DNA. The activity of the clamp loader requires ATP hydrolysis to "close" the clamp around the DNA.
References
Further reading
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.