Chorismate lyase
Protein family From Wikipedia, the free encyclopedia
The enzyme chorismate lyase (EC 4.1.3.40) catalyzes the first step in ubiquinone biosynthesis, the removal of pyruvate from chorismate, to yield 4-hydroxybenzoate in Escherichia coli and other Gram-negative bacteria.[1] It belongs to the family of lyases, specifically the oxo-acid-lyases, which cleave carbon-carbon bonds. The systematic name of this enzyme class is chorismate pyruvate-lyase (4-hydroxybenzoate-forming). Other names in common use include CL, CPL, and UbiC.
| Chorismate lyase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 4.1.3.40 | ||||||||
| CAS no. | 157482-18-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
This enzyme catalyses the chemical reaction:[2]
Activity
Catalytic activity
- This enzyme has an optimum pH at 7.5
Enzymatic activity
Inhibited by:
- Vanillate
- 4-hydroxybenzaldehyde
- 3-carboxylmethylaminmethyl-4-hydroxybenzoic acid
- 4HB - ubiC is inhibited by the product of the reaction, which scientists believe serves as a control mechanism for the pathway
Pathway
The pathway used is called the ubiquinone biosynthesis pathway, it catalyzes the first step in the biosynthesis of ubiquinone in E. coli. Ubiquinone is a lipid-soluble electron-transporting coenzyme. They are essential electron carriers in prokaryotes and are essential in aerobic organisms to achieve ATP synthesis.[4]
Nomenclature
There are several different names for chorismate lyase. It is also called chorismate pyruvate lyase (4-hydroxybenzoate-forming) and it is also abbreviated several different ways: CPL, CL, and ubiC. It is sometimes referred to as ubiC, because that is the gene name. This enzyme belongs to the class lyases; more specifically the ox-acid-lyase or the carbon-carbon-lyases.[5]
Taxonomic lineage:
- bacteria → proteobacteria → gammaproteobacteria → enterobacteriales → enterobacteriaceae → escherichia → Escherichia coli
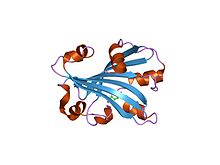
Structure
This enzyme is a monomer. Its secondary structure contains helixes, turns, and beta-strands. It has a mass of 18,777 daltons and its sequence is 165 amino acids long.[5]
Binding sites
- position: 35(M)
- position: 77(R)
- position: 115(L)
Mutagenesis
- position: 91G → A; increases product inhibition by 40%. No effect on substrate affinity.
- position: 156E → K; loss of activity
References
Further reading
Wikiwand - on
Seamless Wikipedia browsing. On steroids.