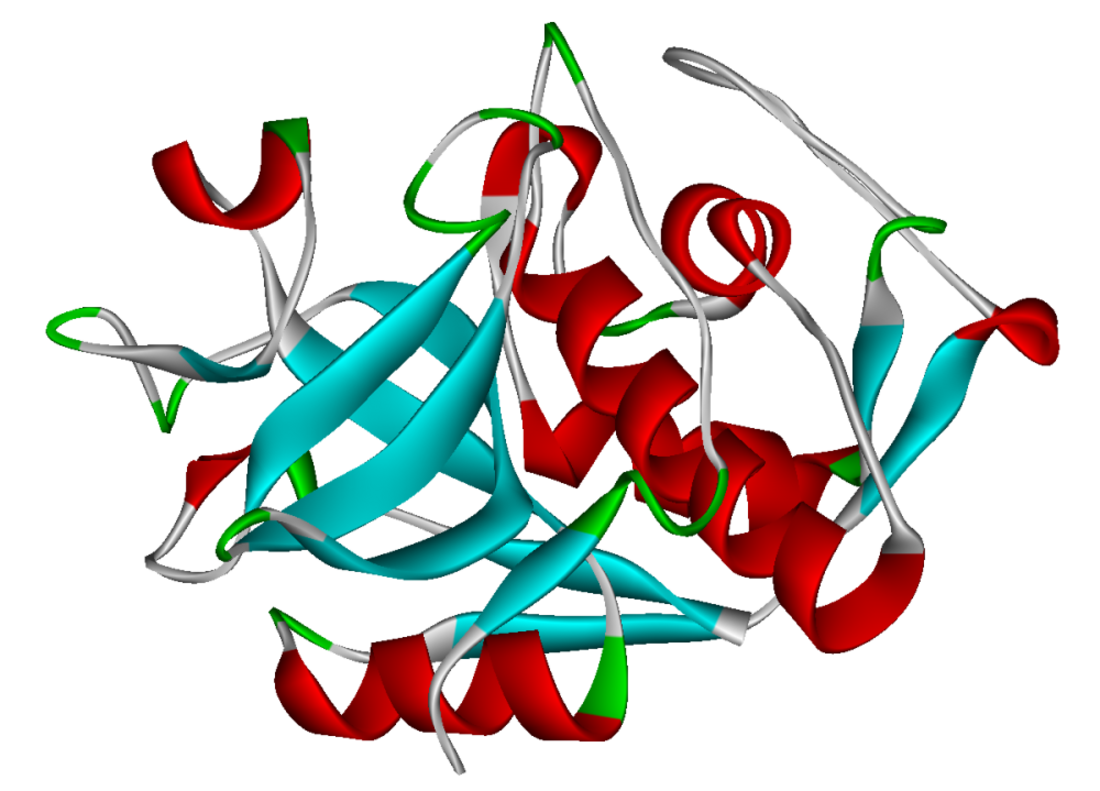
Cathepsin
Family of proteases From Wikipedia, the free encyclopedia
Cathepsins (Ancient Greek kata- "down" and hepsein "boil"; abbreviated CTS) are proteases (enzymes that degrade proteins) found in all animals as well as other organisms. There are approximately a dozen members of this family, which are distinguished by their structure, catalytic mechanism, and which proteins they cleave[citation needed]. Most of the members become activated at the low pH found in lysosomes. Thus, the activity of this family lies almost entirely within those organelles. There are, however, exceptions such as cathepsin K, which works extracellularly after secretion by osteoclasts in bone resorption. Cathepsins have a vital role in mammalian cellular turnover.
Classification
- Cathepsin A (serine protease)
- Cathepsin B (cysteine protease)
- Cathepsin C (cysteine protease)
- Cathepsin D (aspartyl protease)
- Cathepsin E (aspartyl protease)
- Cathepsin F (cysteine proteinase)
- Cathepsin G (serine protease)
- Cathepsin H (cysteine protease)
- Cathepsin K (cysteine protease)
- Cathepsin L1 (cysteine protease)
- Cathepsin L2 (or V) (cysteine protease)
- Cathepsin O (cysteine protease)
- Cathepsin P (mouse cysteine protease)
- Cathepsin Q (rat cysteine protease)
- Cathepsin S (cysteine protease)
- Cathepsin W (cysteine proteinase)
- Cathepsin Z (or X) (cysteine protease)
Clinical significance
Summarize
Perspective
Cathepsins are involved in many physiological processes and have been implicated in a number of human diseases. The cysteine cathepsins have attracted significant research effort as drug targets.[1][2]
- Cancer, Cathepsin D is a mitogen and "it attenuates the anti-tumor immune response of decaying chemokines to inhibit the function of dendritic cells". Cathepsins B and L are involved in matrix degradation and cell invasion.[3]
- Stroke[4]
- Traumatic brain injury[5]
- Alzheimer's disease[6]
- Arthritis[7]
- Ebola, Cathepsin B and to a lesser extent cathepsin L have been found to be necessary for the virus to enter host cells.[8]
- COPD
- Chronic periodontitis
- Pancreatitis
- Several ocular disorders: keratoconus, retinal detachment, age-related macular degeneration, and glaucoma.[9]
Cathepsin A
Deficiencies in this protein are linked to multiple forms of galactosialidosis. The cathepsin A activity in lysates of metastatic lesions of malignant melanoma is significantly higher than in primary focus lysates. Cathepsin A increased in muscles moderately affected by muscular dystrophy and denervating diseases.
Cathepsin B
Cathepsin B may function as a beta-secretase 1, cleaving amyloid precursor protein to produce amyloid beta.[10] Overexpression of the encoded protein, which is a member of the peptidase C1 family, has been associated with esophageal adenocarcinoma and other tumors.[11] Cathepsin B has also been implicated in the progression of various human tumors[3] including ovarian cancer.
Cathepsin D
Cathepsin D (an aspartyl protease) appears to cleave a variety of substrates such as fibronectin and laminin. Unlike some of the other cathepsins, cathepsin D has some protease activity at neutral pH.[12] High levels of this enzyme in tumor cells seems to be associated with greater invasiveness.
Cathepsin K
Cathepsin K is the most potent mammalian collagenase. Cathepsin K is involved in osteoporosis, a disease in which a decrease in bone density causes an increased risk for fracture. Osteoclasts are the bone resorbing cells of the body, and they secrete cathepsin K in order to break down collagen, the major component of the non-mineral protein matrix of the bone.[13] Cathepsin K, among other cathepsins, plays a role in cancer metastasis through the degradation of the extracellular matrix.[14] The genetic knockout for cathepsin S and K in mice with atherosclerosis was shown to reduce the size of atherosclerotic lesions.[15] The expression of cathepsin K in cultured endothelial cells is regulated by shear stress.[16] Cathepsin K has also been shown to play a role in arthritis.[17]
Cathepsin V
Mouse cathepsin L is homologous to human cathepsin V.[18] Mouse cathepsin L has been shown to play a role in adipogenesis and glucose intolerance in mice. Cathepsin L degrades fibronectin, insulin receptor (IR), and insulin-like growth factor 1 receptor (IGF-1R). Cathepsin L-deficient mice were shown to have less adipose tissue, lower serum glucose and insulin levels, more insulin receptor subunits, more glucose transporter (GLUT4) and more fibronectin than wild type controls.[19]
Inhibitors
Five cyclic peptides show inhibitory activity towards human cathepsins L, B, H, and K.[20] Several inhibitors have reached clinical trials, targeting cathepsins K and S as promising therapeutics for osteoporosis, osteoarthritis, and chronic pain. Cathepsin K inhibitors, Relacatib, Balicatib, and Odanacatib, were terminated during clinical trials at phases I, II, and III, respectively, owing to adverse side effects.[21] SAR114137, a Cathepsin S inhibitor, did not progress past phase I for chronic pain. In 2022, STI-1558, a Cathepsin L inhibitor, received FDA clearance to begin phase I studies to treat COVID-19.[22]
Cathepsin zymography
Summarize
Perspective
Zymography is a type of gel electrophoresis that uses a polyacrylamide gel co-polymerized with a substrate in order to detect enzyme activity. Cathepsin zymography separates different cathepsins based on their migration through a polyacrylamide gel co-polymerized with a gelatin substrate. The electrophoresis takes place in non-reducing conditions, and the enzymes are protected from denaturation using leupeptin.[23] After protein concentration is determined, equal amounts of tissue protein are loaded into a gel. The protein is then allowed to migrate through the gel. After electrophoresis, the gel is put into a renaturing buffer in order to return the cathepsins to their native conformation. The gel is then put into an activation buffer of a specific pH and left to incubate overnight at 37 °C. This activation step allows the cathepsins to degrade the gelatin substrate. When the gel is stained using a Coomassie blue stain, areas of the gel still containing gelatin appear blue. The areas of the gel where cathepsins were active appear as white bands. This cathepsin zymography protocol has been used to detect femtomole quantities of mature cathepsin K.[23] The different cathepsins can be identified based on their migration distance due to their molecular weights: cathepsin K (~37 kDa), V (~35 kDa), S (~25kDa), and L (~20 kDa). Cathepsins have specific pH levels at which they have optimum proteolytic activity. Cathepsin K is able to degrade gelatin at pH 7 and 8, but these pH levels do not allow for cathepsins L and V activity. At a pH 4 cathepsin V is active, but cathepsin K is not. Adjusting the pH of the activation buffer can allow for further identification of cathepsin types.[24]
History
Summarize
Perspective
The term cathepsin was coined in 1929 by Richard Willstätter and Eugen Bamann to describe a proteolytic activity of leukocytes and tissues at slightly acidic pH (Willstätter & Bamann (1929) Hoppe-Seylers Z. Physiol. Chemie 180, 127-143). The earliest record of "cathepsin" found in the MEDLINE database (e.g., via PubMed) is from the Journal of Biological Chemistry in 1949.[25] However, references within this article indicate that cathepsins were first identified and named around the turn of the 20th century. Much of this earlier work was done in the laboratory of Max Bergmann, who spent the first several decades of the century defining these proteases.[26]
It is notable that research published in the 1930s (primarily by Bergmann) used the term "catheptic enzymes" to refer to a broad family of proteases that included papain, bromelin, and cathepsin itself.[27] Initial efforts to purify and characterize proteases using hemoglobin transpired at a time when the word "cathepsin" indicated a single enzyme;[28] the existence of multiple, distinct cathepsin family members (e.g. B, H, L) did not appear to be understood at the time. However, by 1937 Bergmann and colleagues began to differentiate cathepsins on the basis of their source in the human body (e.g. liver cathepsin, spleen cathepsin).[26]
References
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.