Alphaproteobacteria
Class of bacteria From Wikipedia, the free encyclopedia
Alphaproteobacteria or α-proteobacteria, also called α-Purple bacteria in earlier literature, is a class of bacteria in the phylum Pseudomonadota (formerly "Proteobacteria").[4] The Magnetococcales and Mariprofundales are considered basal or sister to the Alphaproteobacteria.[5][6] The Alphaproteobacteria are highly diverse and possess few commonalities, but nevertheless share a common ancestor. Like all Proteobacteria, its members are gram-negative, although some of its intracellular parasitic members lack peptidoglycan and are consequently gram variable.[4][2]
| Alphaproteobacteria | |
|---|---|
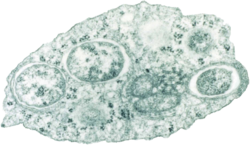 | |
| Transmission electron micrograph of Wolbachia within an insect cell. Credit:Public Library of Science / Scott O'Neill | |
| Scientific classification | |
| Domain: | Bacteria |
| Kingdom: | Pseudomonadati |
| Phylum: | Pseudomonadota |
| Class: | Alphaproteobacteria Garrity et al. 2006 |
| Subclasses[1] and Orders[2] | |
| |
| Synonyms[2] | |
| |
Characteristics
The Alphaproteobacteria are a diverse taxon and comprise several phototrophic genera, several genera metabolising C1-compounds (e.g. Methylobacterium spp.), symbionts of plants (e.g. Rhizobium spp.), endosymbionts of arthropods (Wolbachia) and intracellular pathogens (e.g. Rickettsia). Moreover, the class is sister to the protomitochondrion, the bacterium that was engulfed by the eukaryotic ancestor and gave rise to the mitochondria, which are organelles in eukaryotic cells (see Endosymbiotic theory).[1][7] A species of technological interest is Rhizobium radiobacter (formerly Agrobacterium tumefaciens): scientists often use this species to transfer foreign DNA into plant genomes.[8] Aerobic anoxygenic phototrophic bacteria, such as Pelagibacter ubique, are alphaproteobacteria that are a widely distributed and may constitute over 10% of the open ocean microbial community.
Evolution and genomics
Summarize
Perspective
Several points of disagreement muddy the recovery of the phylogenetic relationships among the Alphaproteobacteria clades from the genomic data. One such point centers on the placement of the Pelagibacterales stemming from the large differences in gene content (e.g. genome streamlining in Pelagibacter ubique) and GC-content between members of several orders.[1] Specifically, certain species within Pelagibacterales, Rickettsiales, and Holosporales possess AT-rich genomes, containing higher-assayed concentrations of adenine-thymine (AT) pairs than guanine-cytosine (GC) base pairs. While it could be a case of convergent evolution resulting in an artefactual clustering,[9][10][11] several studies disagree[1][12][13][14] and no consensus has been reached.
Furthermore, the GC-content of ribosomal RNA, the traditional phylogenetic marker for prokaryotes, does not correlate well with the GC-content of the genome. For example, members of the Holosporales have a much higher ribosomal GC-content than members of the Pelagibacterales and Rickettsiales, though they are more closely related to species with high genomic GC-contents than to members of the latter two orders.[1]
Alphaproteobacteria are divided into three subclasses, Magnetococcidae, Rickettsidae, and Caulobacteridae.[1] The basal group is Magnetococcidae, composed of a large diversity of magnetotactic bacteria only one of which, Magnetococcus marinus, is formally described.[15] The Rickettsidae is composed of the intracellular Rickettsiales and the free-living Pelagibacterales. The Caulobacteridae is composed of the Holosporales, Rhodospirillales, Sphingomonadales, Rhodobacterales, Caulobacterales, Kiloniellales, Kordiimonadales, Parvularculales, and Sneathiellales.
Comparative analyses of the sequenced genomes have revealed many conserved insertion-deletions (indels) in widely distributed proteins and whole proteins (i.e. signature proteins) that are distinctive characteristics of either all Alphaproteobacteria, or their different main orders (viz. Rhizobiales, Rhodobacterales, Rhodospirillales, Rickettsiales, Sphingomonadales and Caulobacterales) and families (viz. Rickettsiaceae, Anaplasmataceae, Rhodospirillaceae, Acetobacteraceae, Bradyrhiozobiaceae, Brucellaceae and Bartonellaceae).
These molecular signatures provide a means to circumscribe the taxonomic groups and to identify and assign new species accurately.[16] Phylogenetic analyses and conserved indels in large numbers of other proteins provide evidence that Alphaproteobacteria have branched off later than most other phyla and classes of Bacteria except Betaproteobacteria and Gammaproteobacteria.[17][18]
Other phylogenetic debates turn on the placement of Magnetococcidae and the protomitochondrion.[19][20] There are some debates for the inclusion of Magnetococcidae in Alphaproteobacteria. For example, an independent proteobacterial class ("Candidatus Etaproteobacteria") for Magnetococcidae has been proposed.[21][22] A recent phylogenomic study suggests the placement of the protomitochondrial clade between Magnetococcidae and all other alphaproteobacterial taxa,[5] which suggests an early divergence of the protomitochondrial lineage from the rest of alphaproteobacteria, except for Magnetococcidae. This phylogeny also suggests that the protomitochondrial lineage does not necessarily have a close relationship to Rickettsidae.
Incertae sedis
Summarize
Perspective
The following taxa have been assigned to the Alphaproteobacteria, but have not been assigned to one or more intervening taxonomic ranks:[23]
- Orders not assigned to a subclass
- Minwuiales Sun et al. 2018
- Genera not assigned to a family
- "Candidatus Anoxipelagibacter" Ruiz-Perez et al. 2021
- "Bilophococcus" Moench 1988
- "Charonomicrobium" Csotonyi et al. 2011
- "Candidatus Endolissoclinum" Kwan et al. 2012
- "Candidatus Endowatersipora" Anderson and Haygood 2007
- "Candidatus Halyseomicrobium" Levantesi et al. 2004
- "Candidatus Halyseosphaera" Kragelund et al. 2006
- "Candidatus Hodgkinia" McCutcheon et al. 2009
- "Candidatus Lariskella" Matsuura et al. 2012
- "Marinosulfonomonas" Holmes et al. 1997
- "Candidatus Mesopelagibacter" Ruiz-Perez et al. 2021
- "Methylosulfonomonas" Holmes et al. 1997
- "Candidatus Monilibacter" Kragelund et al. 2006
- "Nanobacterium" Ciftcioglu et al. 1997
- "Oleomonas" Kanamori et al. 2002
- "Candidatus Paraholospora" Eschbach et al. 2009
- "Candidatus Phycosocius" Tanabe et al. 2015
- "Candidatus Puniceispirillum" Oh et al. 2010
- "Tetracoccus" Blackall et al. 1997
- "Tuberoidobacter" Nikitin 1983[24][25][26]
- Species not assigned to a genus
- Vibrio adaptatus Muir et al. 1990
- Vibrio cyclosites Muir et al. 1990
Phylogeny
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN).[2] The phylogeny is based on whole-genome analysis.[6][a] Subclass names are based on Ferla et al. (2013).[1]
| Bacteria |
| ||||||||||||||||||||||||
Natural genetic transformation
Although only a few studies have been reported on natural genetic transformation in the Alphaproteobacteria, this process has been described in Agrobacterium tumefaciens,[28] Methylobacterium organophilum,[29] and Bradyrhizobium japonicum.[30] Natural genetic transformation is a sexual process involving DNA transfer from one bacterial cell to another through the intervening medium, and the integration of the donor sequence into the recipient genome by homologous recombination.
Notes
- Holosporales and Minwuiales are omitted from this phylogenetic tree.
References
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
