The Alpha variant (B.1.1.7) was[2][3] a SARS-CoV-2 variant of concern. It was estimated to be 40–80% more transmissible than the wild-type SARS-CoV-2 (with most estimates occupying the middle to higher end of this range). Scientists more widely took note of this variant in early December 2020, when a phylogenetic tree showing viral sequences from Kent, United Kingdom looked unusual.[4]
| Alpha | |
 | |
| General details | |
|---|---|
| WHO Designation | Alpha |
| Lineage | B1.1.1.7 |
| First detected | Kent, England |
| Date reported | November 2020 |
| Status | Variant of concern |
| Symptoms | |
| |
| Cases map | |
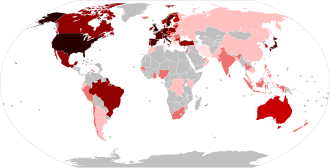 Legend: 10,000+ confirmed sequences
5,000–9,999 confirmed sequences
1,000–4,999 confirmed sequences
500–999 confirmed sequences
100–499 confirmed sequences
2–99 confirmed sequences
1 confirmed sequence
None or no data available
| |
| Major variants | |
The variant began to spread quickly by mid-December, around the same time as infections surged. This increase is thought to be at least partly because of one or more mutations in the virus' spike protein. The variant was also notable for having more mutations than normally seen.[5] By January 2021, more than half of all genomic sequencing of SARS-CoV-2 was carried out in the UK.[6] This gave rise to questions as to how many other important variants were circulating around the world undetected.[7][8]
On 2 February 2021, Public Health England reported that they had detected "[a] limited number of B.1.1.7 VOC-202012/01 genomes with E484K mutations",[9] which they dubbed Variant of Concern 202102/02 (VOC-202102/02).[10] One of the mutations (N501Y) was also present in Beta variant and Gamma variant. On 31 May 2021, the World Health Organization announced that the Variant of Concern would be labelled "Alpha" for use in public communications.[11][12]
The Alpha variant disappeared in late 2021 as a result of competition from even more infectious variants. In March 2022, the World Health Organization changed its designation to "previously circulating variant of concern".
Classification
Names
The variant is known by several names. Outside the UK it is sometimes referred to as the UK variant or British variant or English variant,[13] despite the existence of other, less common, variants first identified in UK, such as Eta variant (lineage B.1.525). Within the UK, it is commonly referred to as the Kent variant after Kent, where the variant was found.[14][15][16]
In scientific use, the variant had originally been named the first Variant Under Investigation in December 2020 (VUI – 202012/01) by Public Health England (PHE),[17][a] but was reclassified to a Variant of Concern (Variant of Concern 202012/01, abbreviated VOC-202012/01) by Meera Chand and her colleagues in a report published by PHE on 21 December 2020.[b] In a report written on behalf of COVID-19 Genomics UK (COG-UK) Consortium, Andrew Rambaut and his co-authors, using the Phylogenetic Assignment of Named Global Outbreak Lineages (pangolin) tool, dubbed it lineage B.1.1.7,[19] while Nextstrain dubbed the variant 20I/501Y.V1.[20]
The name VOC-202102/02 refers to the variant with the E484K mutation (see below).[10]
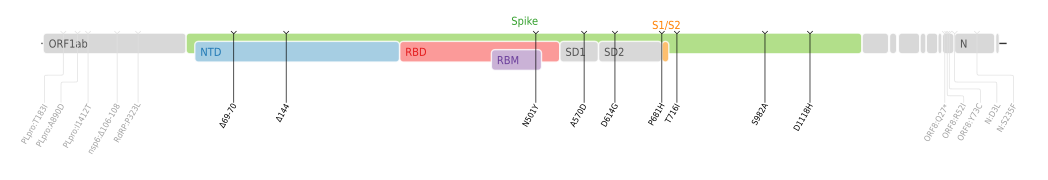
Genetic profile
- Amino acid mutations of SARS-CoV-2 Alpha variant plotted on a genome map of SARS-CoV-2 with a focus on Spike.[21]
| Gene | Nucleotide | Amino acid |
|---|---|---|
| ORF1ab | C3267T | T1001I |
| C5388A | A1708D | |
| T6954C | I2230T | |
| 11288–11296 deletion | SGF 3675–3677 deletion | |
| Spike | 21765–21770 deletion | HV 69–70 deletion |
| 21991–21993 deletion | Y144 deletion | |
| A23063T | N501Y | |
| C23271A | A570D | |
| C23604A | P681H | |
| C23709T | T716I | |
| T24506G | S982A | |
| G24914C | D1118H | |
| ORF8 | C27972T | Q27stop |
| G28048T | R52I | |
| A28111G | Y73C | |
| N | 28280 GAT→CTA | D3L |
| C28977T | S235F | |
| Source: Chand et al., table 1 (p. 5) | ||
Mutations in SARS-CoV-2 are common: over 4,000 mutations have been detected in its spike protein alone, according to the COVID-19 Genomics UK (COG-UK) Consortium.[22]
VOC-202012/01 is defined by 23 mutations: 14 non-synonymous mutations, 3 deletions, and 6 synonymous mutations[23] (i.e., there are 17 mutations that change proteins and six that do not[5]).
Symptoms and signs
Imperial College London investigated over a million people in England while the Alpha variant was dominant and discovered a wide range of further symptoms linked to Covid. "Chills, loss of appetite, headache and muscle aches" were most common in infected people, as well as classic symptoms.[24]
Diagnosis
Several rapid antigen tests for SARS-CoV-2 are in widespread use globally for COVID-19 diagnostics. They are believed to be useful in stopping the chain of transmission of the virus by providing the means to rapidly identify large numbers of cases as part of a mass-testing program. Following the emergence of VOC-202012/01, there was initially concern that rapid tests might not detect it, but Public Health England determined that rapid tests evaluated and used in the United Kingdom detected the variant.[25]
Prevention
By late 2020, several COVID-19 vaccines were being deployed or under development.
However, as further mutations occurred, concerns were raised as to whether vaccine development would need to be altered. SARS-CoV-2 does not mutate as quickly as, for example, influenza viruses, and the new vaccines that had proved effective by the end of 2020 are types that can be adjusted if necessary.[26] As of the end of 2020, German, British, and American health authorities and experts believe that existing vaccines will be as effective against VOC-202012/01 as against previous variants.[27][28]
On 18 December, NERVTAG determined "that there are currently insufficient data to draw any conclusion on... [a]ntigenic escape".[29]
As of 20 December 2020[update], Public Health England confirmed there is "no evidence" to suggest that the new variant would be resistant to the Pfizer–BioNTech vaccine currently being used in the UK's vaccination programme, and that people should still be protected.[30]
E484K mutation
On 2 February 2021, Public Health England reported that they had detected "[a] limited number of B.1.1.7 VOC-202012/01 genomes with E484K mutations",[9] which is also present in the Beta and Gamma variants;[14] a mutation which may reduce vaccine effectiveness.[14] On 9 February 2021, it became known that some 76 cases with the E484K mutation had been detected, principally in Bristol, but with a genomically distinct group in Liverpool also carrying the mutation.[31] A week later a Research and analysis report from PHE gave a total of 77 confirmed and probable cases involving the E484K mutation across the UK, in two variants, VUI-202102/01 and VOC-202102/02, the latter described as 'B.1.1.7 with E484K'.[10]
On 5 March 2021, it was reported that a B.1.1.7 lineage with the E484K mutation has been detected in two US patients (in Oregon and New York states). Researchers thought that the E484K mutation in the Oregon variant arose independently.[32][33]
Characteristics
Transmissibility
The transmissibility of the Alpha variant (lineage B.1.1.7) had generally been found to be substantially higher than that of pre-existing SARS-CoV-2 variants. The variant was discovered by a team of scientists at COG-UK whose initial results found transmissibility was 70% (50-100%) higher.[16][34] A study by the Centre for the Mathematical Modelling of Infectious Diseases at the London School of Hygiene & Tropical Medicine reported that the variant was 43 to 90% (range of 95% credible intervals, 38 to 130%) more transmissible than pre-existing variants in the United Kingdom, depending on the method used to assess increases in transmissibility. Similar increases in the transmissibility of lineage B.1.1.7 were measured in Denmark, Switzerland, and the United States.[35] Furthermore, a simple model to account for the rapid rise of lineage B.1.1.7 in several countries and the world found that the variant is 50% more transmissible than the local wild type in these three countries and across the world as whole.[36] Another study concluded that it was 75% (70%–80%) more transmissible in the UK between October and November 2020.[37] A later study suggested that these earlier estimates overestimated the transmissibility of the variant and that the transmissibility increase was on the lower ends of these ranges.[38][39]
The Dutch Ministry of Health, Welfare and Sport calculated, based on genome sequencing of positive cases, each week the transmissibility rate of the variant compared to the local wildtype, and found it to fluctuate between 28%-47% higher during the first six weeks of 2021.[40] The Danish Statens Serum Institut in comparison calculated it to be 55% (48%–62%) more transmissible in Denmark based upon the observed development of its relative frequency from 4 January to 12 February 2021.[41] The Institute of Social and Preventive Medicine (ISPM) under University of Bern, calculated the transmissibility of lineage B.1.1.7 based on the weekly development of its observed fraction of all Covid-19 positives during the entire pandemic, and found for 95% confidence intervals under the assumption of a wildtype reproduction number Rw≈1 and an exponentially generation time of 5.2 days, that transmissibility was: 52% (45%–60%) higher when compared to the wildtype in Denmark and 51% (42%-60%) higher when compared to the wildtype in Switzerland.[42]
On 18 December 2020—early on in the risk assessment of the variant—the UK scientific advisory body New and Emerging Respiratory Virus Threats Advisory Group (NERVTAG) concluded that they had moderate confidence that VOC-202012/01 was substantially more transmissible than other variants, but that there were insufficient data to reach any conclusion on underlying mechanism of increased transmissibility (e.g. increased viral load, tissue distribution of virus replication, serial interval etc.), the age distribution of cases, or disease severity.[29] Data seen by NERVTAG indicated that the relative reproduction number ("multiplicative advantage") was determined to be 1.74—i.e., the variant is 74% more transmissible—assuming a 6.5-day generational interval. It was demonstrated that the variant grew fast exponentially with respect to the other variants.[43][44][45] The variant out-competed the ancestral variant by a factor of every two weeks. Another group came to similar conclusions, generating a replicative advantage, independent of "protective measures", of 2.24 per generation of 6.73 days, out-competing the ancestral variant by every two weeks.[46] Similarly, in Ireland, the variant—as indicated by the missing S gene[c] detection (S-gene target failure [SGTF]), which historically was rare—went from 16.3% to 46.3% of cases in two weeks. This demonstrates, based on the statistics of 116 positive samples, that the variant had a relative higher growth by a factor of , when compared to the average growth for all other variants by the end of this two week period.[48] The variant became the dominant variant in London, East of England and the South East from low levels in one to two months. A surge of SARS-CoV-2 infections around the start of the new year is seen[by whom?] as being the result of the elevated transmissibility of the variant, while the other variants were in decline.[49][50][51]
One of the most important changes in lineage B.1.1.7 seems to be N501Y,[22] a change from asparagine (N) to tyrosine (Y) in amino-acid position 501.[52] This is because of its position inside the spike protein's receptor-binding domain (RBD)—more specifically inside the receptor-binding motif (RBM),[53] a part of the RBD[54]—which binds human ACE2.[55] Mutations in the RBD can change antibody recognition and ACE2 binding specificity[55] and lead to the virus becoming more infectious;.[22] Chand et al. concluded that "[i]t is highly likely that N501Y affects the receptor-binding affinity of the spike protein, and it is possible that this mutation alone or in combination with the deletion at 69/70 in the N-terminal domain (NTD) is enhancing the transmissibility of the virus".[56] In early 2021 a peer-reviewed paper found that the mentioned HV 69–70 deletion in vitro "appeared to have two-fold higher infectivity over a single round of infection compared to [wild-type SARS-CoV-2]" in lentiviral SARS-CoV-2 pseudoviruses.[57]
Using In Silico approach, Shahhosseini et al. demonstrated that the Y501 mutation in lineage B.1.1.7 forms a shorter H-bond (length of 2.94 Å) than its counterpart in the wild type (WT) variant residue N501, indicating that in lineage B.1.1.7 the RBD and hACE2 have a stable interaction. Furthermore, the Y501 mutation in lineage B.1.1.7 contributes more negatively to Binding Free Energy (BFE) (-7.18 kcal/mol) than its counterpart in the WT variant residue N501 (-2.92 kcal/mol). As a result of combining BFE and molecular interaction results, the N501Y mutation in RBD strengthens binding affinity of SARS-CoV-2 RBD to hACE2.[58]
In a detailed affinity and kinetic analysis of the interaction between the Spike RBD and ACE2, the N501Y mutation was found to significantly enhance the binding affinity between the RBD and ACE2 by approximately 10-fold, resulting from a 1.8-fold increase in the association rate constant (kon) and a 7-fold decrease in the dissociation rate constant (koff).[59]
Virulence
Matched cohort studies of the Alpha variant (lineage B.1.1.7) found higher mortality rate than earlier circulating variants overall,[60][61] but not in hospitalised patients.[62] An ecological study found no difference in mortality overall.[63]
Initially, NERVTAG said on 18 December 2020 that there were insufficient data to reach a conclusion regarding disease severity. At prime minister Boris Johnson's briefing the following day, officials said that there was "no evidence" as of that date that the variant caused higher mortality or was affected differently by vaccines and treatments;[64] Vivek Murthy agreed with this.[65] Susan Hopkins, the joint medical adviser for the NHS Test and Trace and Public Health England (PHE), declared in mid-December 2020: "There is currently no evidence that this strain causes more severe illness, although it is being detected in a wide geography, especially where there are increased cases being detected."[22] Around a month later, however—on 22 January 2021—Johnson said that "there is some evidence that the new variant [VOC-202012/01]... may be associated with a higher degree of mortality", though Sir Patrick Vallance, the government's Chief Scientific Advisor, stressed that there is not yet enough evidence to be fully certain of this.[66]
In a paper analysing twelve different studies on lineage B.1.1.7 death rate relative to other lineages, it was found to have a higher death rate (71% according to LSHTM, 70% according to University of Exeter, 65% according to Public Health England, and 36% according to Imperial College London), and NERVTAG concluded: "Based on these analyses, it is likely that infection with VOC B.1.1.7 is associated with an increased risk of hospitalisation and death compared to infection with non-VOC viruses".[67] Results of the death studies were associated with some high uncertainty and confidence intervals, because of a limited sample size related to the fact that UK only analysed the VOC status for 8% of all COVID-19 deaths.[68]
A UK case-control study for 54,906 participants, testing positive for SARS-CoV-2 between 1 October 2020 and 29 January 2021 in the community setting (not including vulnerable persons from care centres and other public institutions), reported that patients infected with the Alpha variant (VOC 202012/1) had a hazard ratio for death within 28 days of testing of 1.64 (95% confidence interval 1.32-2.04), as compared with matched patients positive for other variants of SARS-CoV-2.[69] Also in the UK, a survival analysis of 1,146,534 participants testing positive for SARS-CoV-2 between 1 November 2020 and 14 February 2021, including individuals in the community and in care and nursing homes, found a hazard ratio of 1.61 (95% confidence interval 1.42–1.82) for death within 28 days of testing among individuals infected with lineage B.1.1.7; no significant differences in the increased hazard of death associated with lineage B.1.1.7 were found among individuals differing in age, sex, ethnicity, deprivation level, or place of residence.[61] Both studies adjusted for varying COVID-19 mortality by geographical region and over time, correcting for potential biases due to differences in testing rates or differences in the availability of hospital services over time and space.[citation needed]
A Danish study found people infected by lineage B.1.1.7 to be 64% (32%–104%) more likely to get admitted to hospitals compared with people infected by another lineage.[70]
Genetic sequencing of VOC-202012/01 has shown a Q27stop mutation which "truncates the ORF8 protein or renders it inactive".[19] An earlier study of SARS-CoV-2 variants which deleted the ORF8 gene noted that they "have been associated to milder symptoms and better disease outcome".[71] The study also noted that "SARS-CoV-2 ORF8 is an immunoglobulin (Ig)–like protein that modulates pathogenesis", that "SARS-CoV-2 ORF8 mediates major histocompatibility complex I (MHC-I) degradation", and that "SARS-CoV-2 ORF8 suppresses type I interferon (IFN)–mediated antiviral response".[71]
Referring to amino-acid position 501 inside the spike protein, Chand et al. concluded that "it is possible that variants at this position affect the efficacy of neutralisation of virus",[56] but noted that "[t]here is currently no neutralisation data on N501Y available from polyclonal sera from natural infection".[56] The HV 69–70 deletion has, however, been discovered "in viruses that eluded the immune response in some immunocompromised patients",[72] and has also been found "in association with other RBD changes".[56]
Epidemiology


Cases of the Alpha variant (lineage B.1.1.7) were estimated to be under-reported by most countries as the most commonly used tests do not distinguish between this variant and other SARS-CoV-2 variants, and as many SARS-CoV-2 infections are not detected at all. RNA sequencing is required for detection of this variant,[82] although RT-PCR test for specific variants[d] can be used as a proxy test for Alpha — or as a supplementing first-screening test before conducting the whole-genome sequencing.[83][75]
As of 23 March, the Alpha variant had been detected in 125 out of 241 sovereign states and dependencies (or 104 out of 194 WHO member countries),[e] of which some had reported existence of community transmission while others so far only found travel related cases.[87] As of 16 March, it had become the dominant COVID-19 variant for 21 countries: United Kingdom (week 52), Ireland (week 2), Bulgaria (week 4), Slovakia (week 5), Israel (week 5), Luxembourg (week 5), Portugal (week 6), Denmark (week 7), Netherlands (week 7), Norway (week 7), Italy (week 7), Belgium (week 7), France (week 8), Austria (week 8), Switzerland (week 8), Liechtenstein (week 9), Germany (week 9), Sweden (week 9), Spain (week 9), Malta (week 10) and Poland (week 11). The emergence and the fast spreading of the new variant has been detected in Lebanon and a relationship noted between SARS-CoV-2 transmission intensity and the frequency of the new variant during the first twelve days of January.[88] By February, Alpha became the dominant variant in Lebanon.[89]
Spread in UK
The first case was likely in mid-September 2020 in London or Kent, United Kingdom.[90] The variant was sequenced in September.[91] As of 13 December 2020, 1,108 cases with this variant had been identified in the UK in nearly 60 different local authorities. These cases were predominantly in the south east of England. The variant has also been identified in Wales and Scotland.[92] By November, around a quarter of cases in the COVID-19 pandemic in London were being caused by the new variant, and by December, that was a third.[93] In mid-December, it was estimated that almost 60 percent of cases in London involved Alpha.[94] By 25 January 2021, the number of confirmed and probable UK cases had grown to 28,122.[95]
Spread in Europe
The variant became dominant for:
- South East England in week 48, the last week of November 2020.[43]
- England in week 51 of 2020.[96]
- United Kingdom in week 52 of 2020.[77]
- Scotland and Northern Ireland in week 1 of 2021.[79]
- Wales in week 2 of 2021.[79]
- Ireland in week 2 of 2021.[48]
In Bulgaria, genome sequencing found the variant to be dominant with 52.1% in week 4, followed by 73.4% in week 9.[97]

Also in Slovakia, a RT-PCR Multiplex DX test capable of detecting the 2 deletions specific for lineage B.1.1.7 (ΔH69/ΔV70 and ΔY144),[98] first found the variant nationwide in 74% of cases on 3 February (week 5), followed by 72% of cases on 15 February (week 7), and it then grew to 90% of cases on 3 March (week 9).[99] The same test found earlier on 8 January prevalence of the variant at a rate of 36% in the Michalovce District and 29% in Nitra.[100]
In Israel, the variant was first time detected by genome sequencing 23 December 2020.[101] Leumit Health Care Services however analysed with the proxy test RT-PCR (SGTF) and found the variant at a rate of 3‑4% on 15 December.[102] The national Ministry of Health estimated based on genome sequencing, that the prevalence of the variant became dominant (70%) on 6 February[103] followed by 90% on 16 February.[104]
In Luxembourg, a weekly genome sequencing revealed that the variant grew from 0.3% (week 51) to be dominant with a share of 53.0% in week 5, although results might not be fully representative due to the fact that no correction occurred from potential targeting bias from contact tracing, airport travellers and local outbreaks. Genome sequencing of a population representative randomized test pool - with no target bias - was conducted since week 6, and it confirmed the dominant status of the variant at a rate of 54.1% (week 6) growing to 62.8% (week 9), while the competing Beta variant (lineage B.1.351) was found to be at 18.5% in week 9.[105]
In Denmark the variant grew from 0.3% (week 46 of 2020) to become dominant with 65.9% (week 7 of 2021), and it grew further to 92.7% (week 10); with the regional prevalence ranging from 87.3% in the North Jutland Region to 96.1% in the Central Denmark Region.[106] The observed growth of the relative variant share, was in full accordance with the earlier modelled forecast,[41] that had predicted dominance (over 50%) around mid-February and a prevalence of around 80% of the total circulating variants by early March.[107] In comparison, the genome sequencing only found the competing Beta variant in 0.4% of the positive cases (9 times out of 2315 tests) in week 10.[108][109]
In the Netherlands, a randomly conducted genome sequencing found that the variant grew from 1.3% of cases in week 49 to a dominant share of 61.3% in week 7, followed by 82.0% in week 9; while the competing Beta variant in comparison was found to be at 3.0% in week 9.[74] In Amsterdam, the Alpha variant (lineage B.1.1.7) grew from 5.2% (week 52) to 54.5% (week 6).[110]
In Norway, the variant was found by genome sequencing to grow from 5.7% (week 1) into dominance by a 58.4% (week 7), followed by 65.0% (week 8).[111] Another large survey comprising results of both genome sequencing and PCR proxy tests, with a sample seize of more than 1000 tests per week (since week 4), at the same time found that the variant grew from 2.0% (week 48) into dominance by 60.0% (week 7), followed by 72.7% in week 10 - while only 2.2% of cases in comparison were found to be of the Beta variant.[112] The variant regionally had its highest share in the county of Oslo and Viken,[113] growing from 18% to 90% of analysed samples in Oslo from 20 January to 23 February (although with the data-corrected estimate a bit lower at 50-70% on 23 February); while growing from 21% to 80% of analysed samples in Viken from 25 January to 23 February (although with the data-corrected estimate a bit lower at 50% on 23 February).[114] For the period 15 February to 14 March, the combined survey of genome sequencing and PCR proxy tests, also found the Alpha variant was at a dominant rate over 50% for 8 out of 11 regions, with its highest rate (82%) found for Oslo; while the region Nordland was different from all other regions by having only 6% cases of Alpha along with a dominant 88% of cases represented by the Beta variant.[112]
In Portugal, the variant represented according to a national genome sequencing survey: 16.0% of the Covid-19 infections during 10–19 January (week 2),[115] followed by a dominant 58.2% in week 6.[116] A national RT-PCR proxy test based on SGTF and SGTL observations, found the variant at a rate of 33.5% in week 4, but observed afterwards a decelerating pace for the weekly rise of the variant share (reason unknown), and according to this study it only became dominant by 50.5% (91.8% of 55.0% SGTFL) in week 8, followed by 64.3% (91.8% of 70% SGTFL) in week 10.[75]
In Italy, the Alpha variant accounted for 17.8% of cases nationwide on 4–5 February (week 5),[117] followed by 54.0% on 18 February (week 7). The regional prevalence for week 7 ranged from 0% in Aosta Valley (although only one sample was tested) to 93.3% in Molise. In week 7, the competing Gamma variant had a prevalence of 4.3% (ranging between 0%-36.2% regionally) and the South African variant a prevalence of 0.4% (ranging between 0%-2.9% regionally).[118]
In Switzerland, a nationwide weekly genome sequencing survey found that the Alpha variant grew from 0.05% (week 51) to a dominant 58.2% of cases in week 8, followed by 71.1% in week 9.[76] This was in full accordance with a model from 9 February that had forecast dominance around mid-February.[119] In comparison, the competing Beta variant was only found nationwide in 1.0% of the positive cases in week 9.[76]
In Belgium, genome sequencing of samples selected randomly after excluding all samples from active targeted testing related to local outbreaks or travels (creating a statistical representative national sample with a seize equal to 4.4% of all COVID-19 positive tests), found that the Alpha variant share grew from 7.1% in week 1 to a dominant 51.5% in week 7, followed by 79.3% in week 10.[120] The variant was first time detected by targeted genome sequencing in week 49, but due to a small sample seize (not being random, and less than 100 tests per week) then no reliable variant share data could be determined before week 1.[121] The proxy test for the variant (RT-PCR SGTF) was also conducted for a sample seize equal to 25.8% of all COVID-19 positive tests, and found a dominant 54.8% SGTF rate for week 10. In comparison, the competing Beta variant was found to be at 3.6% and the Gamma variant had a prevalence of 1.8% in week 10.[120]
In France, scientists accurately forecast the Alpha variant (VOC-202012/01) would likely become dominant nationwide around week 8–11 of 2021.[122] A nationwide survey of randomly selected positive COVID-19 samples first analysed by a RT-PCR screening test and subsequently confirmed by genome sequencing, revealed that the variant grew from a share of 3.3% (388/11916) on 7–8 January (week 1)[123] to 13.3% (475/3561) on 27 January (week 4),[85] followed by 44.3% (273/615) on 16 February (week 7).[85] In week 8, the variant was found to have a dominant share of 56.4% (758/1345) according to the interpretable results of a weekly genome sequencing survey comprising 0.9% of all COVID-19 positive tests, or 59.5% according to a variant-specific RT-PCR survey testing 54% of all the COVID-19 positive tests.[124] In week 10, the variant was found to have a share of 71.9% according to a variant-specific RT-PCR survey testing 56.9% of all the COVID-19 positive tests. The spread of the variant differed regionally for the 96 departments located in Metropolitan France for week 10, with 91 departments over 50% and 5 departments with 30%-50% (of which the Moselle department in particular was notable due to finding a high 38.3% rate of the competing Beta variant, that - despite having declined from a dominant 54.4% value in week 8 - still was significantly above the 5.0% national average for this specific variant).[125]
In Austria, Agentur für Gesundheit und Ernährungssicherheit (AGES) collected data from N501Y RT-PCR specific tests combined with subsequent genome sequencing analysis, and found that the variant grew from 7.2% (week 1) to a dominant 61.3% (week 8), followed by 61.2% in week 9 and 48.3% in week 10. If all N501Y positive tests had been analysed further by genome sequencing, then these listed shares could have been even higher, for example they could have been as mcuh as: 2.4% higher for week 1, 23.0% higher for week 7, 4.0% higher for week 8, 6.8% higher for week 9 and 25.4% higher for week 10. The competing Beta variant was only found nationwide in 0.3% of the positive cases in week 10, and for the region Tyrol - where it had been most prevalent - its share declined from 24.5% in week 4 to just 1.9% in week 10. Regionally Alpha was found to be dominant with over 50% for 7 out of 9 regions, with the only two exceptions being Tyrol and Vorarlberg.[126]
In Germany, the largest and probably most representative national survey published by the Robert Koch Institute (entitled RKI-Testzahlerfassung), determined the share of circulating COVID-19 variants for the latest week by analysing 53,272 COVID-19 positive samples either by genome sequencing or RT-PCR proxy tests, with data collected on a voluntary basis from 84 laboratories from the university / research / clinical / outpatient sector - spread evenly across the nation. The survey did not utilize data weights or data selection criteria to ensure existence of geographical representativity, but might still be regarded as somewhat representative for the national average due to its big sample seize. According to the RKI-Testzahlerfassung survey, the variant grew from a share of 2.0% (week 2) to a dominant share of 54.5% (week 9), followed by 63.5% in week 10. In comparison, the competing Beta variant was only found nationwide in 0.9% of the positive cases in week 10.[127]
In Malta, the variant was first time detected by genome sequencing on 30 December 2020,[128] and represented 8% of the positive cases in week 7.[129] A new RT-PCR variant specific test was introduced for the surveillance,[130] where the first results reported on 10 March revealed the variant now represented 61% of cases nationwide.[131]
In Sweden, the national authorities initially expected the variant would become dominant around week 12–14 under the assumption of 50% increased transmissibility compared to the original virus.[132] In average, the variant share was found growing from 10.8% (week 4) to 36.9% (week 7) across five of its southern regions (Skåne, Västra Götaland, Västmanland, Gävleborg and Örebro).[133] For week 7, the share of the variant was for the first time also calculated to 30.4% as the overall average for 19 out of 21 Swedish regions (ranging from 3.3% in Blekinge to 45% in Gävleborg). For week 9, the share of the variant was calculated to a dominant 56.4% as the overall average for the 19 regions (ranging from 16% in Kronoberg to 72% in Gävleborg). For week 10, the share of the variant was calculated to have increased further to 71.3% as the overall average for the 19 regions (ranging from 40% in Kronoberg to 84% in Jönköping). Although no geographical weights were applied to ensure geographical representativity for the calculated average for the 19 regions, the overall sample seize of 12,417 variant tests represented 43.1% of all COVID-19 positive PCR-tests for week 10, inferring that the result of the survey might be close to represent the actual true average for the nation as a whole.[86]
In the microstate Liechtenstein, the first case of Alpha was detected on 19 December 2020. During the entire pandemic, 67 VOC-N501Y PCR-positive cases were detected comprising 58 cases of Alpha, 1 of Beta (detected 1 February) and 8 un-identified N501Y cases (from 10 Dec. to 22 Feb.), as of 18 March 2021. The weekly average of each days calculated 7-day rolling average for the share of all detected VOC-N501Y cases (which is a good proxy for Alpha in Liechtenstein), was found to be 10.9% in week 5, followed by a dominant 52.2% in week 9 (where 8 out of 15 tests were found to be Alpha); and grew further to 73.4% in week 10 (where 6 out of 9 tests were found to be Alpha).[134]
In Spain, the variant share was estimated nationwide to be 5%-10% of cases on 29 January,[135] followed by 20%-25% of cases on 18 February[136][137] and 25%-30% of cases on 22 February,[138] and estimated to be dominant with over 50% as of 3 March (week 9).[139] As of week 10, the prevalence of the variant ranged from 18.3% to 97.0% for the 17 regions, with all but two regions having a dominant rate above 50%:[140]
- No data reported by Extremadura.
- 18.3% for week 10 in Aragon.
- 27.0% for week 7 in Canary Islands.
- 51.3% for week 8 in Castile and León.
- 52.2% for week 10 in Valencia.
- 52.8% for week 10 in Castilla–La Mancha.
- 53.3% for week 4 in Galicia.
- 59.5% for week 9 in La Rioja.
- 59.7% for week 10 in Madrid.
- 61.6% for week 11 in Andalusia.
- 76.1% for week 10 in Balearic Islands.
- 76.7% for week 10 in Murcia.
- 77.4% for week 10 in Basque Country.
- 77.8% for week 10 in Navarre.
- 83.1% for week 10 in Cantabria.
- 84.6% for week 9 in Catalonia.
- 97.0% for week 10 in Asturias.
In Poland, a national survey among infected teachers led to experts estimating that the variant share was between 5% and 10% nationwide as of 11 February,[141] while ECDC reported it to be 9% as of 15 February.[142] According to Health minister Adam Niedzielski, the variant was found at a rate of 5% in the first studies from the second half of January, and then it increased by ten percentage points every ten days, until it became dominant nationwide by a 52% rate on 16 March (week 11).[143][144] Regionally, the variant had already exceeded 70% for Warmian-Masurian and Pomeranian on 22–23 February (week 8),[145] and even reached 90% for the Greater Poland Voivodeship on 17 March (week 11).[146]
In Finland, no statistical representative national survey had been conducted as of February 2021, as the national genome sequencing mainly targeted further analysis of COVID-19 positive samples from travellers and local outbreak clusters.[147] Helsinky University Hospital (HUS), operating in the Helsinki and Uusimaa Hospital District, found the variant in 10% of all samples collected randomly during a few days ahead of 14 February in the capitol region (also known as the Helsinky constituency and Uusimaa constituency). For the capitol region, the variant was modelled to become dominant (over 50%) in the second half of March (week 11-13).[148]
In Iceland, the national authorities implemented a strict 3 test and quarantine regime for all persons entering the country from abroad, that so far successfully managed to prevent new infectious VOCs from gaining a foothold in the country. As of 4 March, a total of 90 travellers had tested positive for the Alpha variant at Iceland's borders and the 20 additional domestic cases were all closely connected to the border cases, with no cases related to community transmission.[149]
Spread in North America
In the United States, the variant first appeared late November 2020,[150] grew from 1.2% in late January and became predominant around the end of March.[151][152]The spread of the variant was primarily concentrated in the upper Midwest of the country, especially the states of Michigan and Minnesota, but it failed to spread in high concentrations to other parts of the country.[153]
In Canada, the variant first appeared in Ontario late December 2020.[154] By 13 February, it had spread to all ten provinces.[155] Testing and confirmation of the Alpha variant (lineage B.1.1.7) in COVID-19-positive samples has been inconsistent across the country.[156] On 3 February, the province of Alberta was the first to screen all COVID-19 positive samples for variants of concern.[156][157] As of 23 March, the Alpha variant had been detected in 5812 cases, and was most prevalent in the province of Alberta.[158] In Ontario, a combined RT-PCR (N501Y) and genome sequencing study found that all VOCs represented 4.4% of all COVID-19 positives on 20 January (week 3), and that Alpha comprised 99% of all those VOCs.[159] Public Health Ontario laboratories found the variant in close to 7% of all COVID-19 positives in week 5, representing 97% (309/319) of all detected VOCs.[160] Alpha became the dominant variant on or around 16 March; 53% of all positive cases were VOCs, and it was presumed 97% of VOCs were Alpha.[161] In Quebec, where the variant was also widespread, it was expected to become dominant by late March or in April.[162]
Development
| Development of the Alpha variant (lineage B.1.1.7) (share of analysed SARS-CoV-2–positive tests in a given week) | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Country | Test | 2020 Week 42 | Week 43 | Week 44 | Week 45 | Week 46 | Week 47 | Week 48 | Week 49 | Week 50 | Week 51 | Week 52 | Week 53 | 2021 Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 | Week 8 | Week 9 | Week 10 |
| United Kingdom | Seq.[77] | 0.05% | 0.35% | 1.0% | 2.7% | 6.3% | 10.2% | 10.2% | 13.9% | 32.9% | 45.7% | 51.3% | 70.6% | 74.1% | 78.6% | 86.0% | 89.1% | – | – | – | – | – | – |
| England | SGTF[163]*Seq.%[164] |
0.07% (0.02%) |
0.4% (0.5%) |
0.9% (1.2%) |
2.9% (1.5%) |
5.5% (5.3%) |
9.1% (7.8%) |
18.0% (11.4%) |
32.2% (22.1%) |
51.8% (38.1%) |
63.7% (61.8%) |
72.4% (62.1%) |
77.9% (78.4%) |
82.1% (76.7%) |
87.5% (79.7%) |
90.6% (–) |
93.8% (–) |
95.7% (–) |
97.4% (–) |
98.1% (–) |
98.6% (–) |
98.7% (–) |
– (–) |
| Northern Ireland | SGTF rawdata[f] SGTF model[f] |
N/A | N/A | N/A | N/A | N/A | 17.9% (m:14.0%) |
7.4% (m:14.9%) |
37.5% (m:16.3%) |
23.0% (m:18.5%) |
50.0% (m:36.1%) |
25.0% (m:42.1%) |
58.8% (m:44.7%) |
45.2% (m:52.7%) |
71.4% (m:67.0%) |
78.2% (m:74.0%) |
72.4% (m:81.4%) |
95.7% (m:86.5%) |
88.9% (m:90.5%) |
92.2% (m:92.4%) |
100.0% (m:93.0%) |
86.2% (m:92.6%) |
100.0% (m:91.1%) |
| Scotland | SGTF rawdata[f] SGTF model[f] |
N/A | N/A | N/A | N/A | N/A | 9.5% (m:5.8%) |
9.7% (m:6.7%) |
10.4% (m:7.7%) |
11.7% (m:10.2%) |
51.4% (m:26.2%) |
40.0% (m:39.5%) |
35.8% (m:43.9%) |
64.1% (m:49.8%) |
67.2% (m:64.2%) |
63.3% (m:65.0%) |
69.2% (m:70.0%) |
70.6% (m:77.3%) |
93.9% (m:84.7%) |
82.0% (m:90.0%) |
100.0% (m:93.7%) |
93.0% (m:95.8%) |
100.0% (m:96.9%) |
| Wales | SGTF rawdata[f] SGTF model[f] |
N/A | N/A | N/A | N/A | N/A | 27.9% (m:18.1%) |
8.3% (m:16.0%) |
10.7% (m:13.3%) |
26.3% (m:12.1%) |
13.4% (m:15.0%) |
15.9% (m:14.7%) |
19.8% (m:20.4%) |
54.4% (m:34.8%) |
61.5% (m:59.6%) |
69.0% (m:65.0%) |
68.1% (m:68.7%) |
77.0% (m:73.4%) |
71.8% (m:76.6%) |
75.8% (m:78.5%) |
87.9% (m:78.9%) |
80.0% (m:77.2%) |
69.9% (m:73.3%) |
| Ireland | SGTF rawdata[48] | – | – | – | 1.9% (few data) |
0.0% (few data) |
0.0% (few data) |
6.1% (few data) |
2.2% (few data) |
1.6% (few data) |
7.5% | 16.3% | 26.2% | 46.3% | 57.7% | 69.5% | 75.0% | 90.1% | 88.6% | 90.8% | – | – | – |
| Bulgaria | Seq.[97] | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 52.1% | – | 70.8% | – | – | 73.4% | – |
| Israel | SGTF w.51+1 Seq. after w.1 |
– | – | – | – | – | – | – | – | – | 3–4% (Dec.15)[102] |
– | – | 10–20% (Jan.5)[102] |
10–20% (Jan.11)[167] |
30–40% (Jan.19)[168] |
40–50% (Jan.25)[169] |
~70% (Feb.2)[103] |
~80% (Feb.9)[170] |
~90% (Feb.16)[104] |
– | – | – |
| Slovakia | PCR (SGTF+ΔY144) [99] | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ~74% (Feb.3) |
– | ~72% (Feb.15) |
– | ~90% (Mar.3) |
– |
| Luxembourg | Seq.[105] | – | – | – | – | – | – | – | – | – | 0.3% | 0.9% | 4.2% | 8.7% | 15.5% | 16.9% | 36.2% | 53.0% | 54.1% | 53.8% | 63.7% | 62.8% | – |
| Denmark | Seq.[106] | – | – | – | – | 0.3% | 0.2% | 0.4% | 0.4% | 0.4% | 0.8% | 1.8% | 1.9% | 3.5% | 7.0% | 13.1% | 19.6% | 29.6% | 47.0% | 65.9% | 75.3% | 85.1% | 92.7% |
| Netherlands | Seq.[74] | – | – | – | – | – | – | – | 1.3% | 0.8% | 0.5% | 2.1% | 4.4% | 9.6% | 15.9% | 22.7% | 24.7% | 31.0.% | 40.3% | 61.3% | 75.2% | 82.0% | – |
| Norway | PCR proxy and Seq.[112] Seq.[171][111] |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
0.0% (–) |
2.0% (–) |
0.0% (0.0%) |
4.9% (0.0%) |
8.9% (7.2%) |
6.0% (2.9%) |
14.9% (1.3%) |
18.9% (5.7%) |
19.5% (11.6%) |
15.2% (9.3%) |
19.8% (20.1%) |
34.1% (30.3%) |
43.6% (40.8%) |
60.0% (58.4%) |
71.8% (65.0%) |
76.7% (–) |
72.7% (–) |
| Italy | Seq.%[117][118] | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 17.8% | – | 54.0% | – | – | – |
| France | PCR proxy Seq. SGTF*Seq.% |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (–) |
– (–) (3.3%)[123] |
– (–) (–) |
– (–) (–) |
– (–) (13.3%)[85] |
– (–) (–) |
36.1%[172] (–) (–) |
49.3%[173] (45.8%)[173] (44.3%)[85] |
59.5%[124] (56.4%)[124] (–) |
65.8%[85] (69.0%)[125] (–) |
71.9%[125] (–) (–) |
| Switzerland | Seq.[76] | – | – | – | – | – | – | – | – | – | 0.05% | 0.4% | 0.8% | 2.6% | 4.9% | 9.2% | 16.5% | 26.0% | 32.4% | 47.0% | 58.2% | 71.1% | – |
| Austria | PCR(N501Y)*Seq.%[126] Seq.B117+unseq.N501Y[126] |
– | – | – | – | – | – | – | – | – | – | – | – | 7.2%[g] (9.6%) |
17.4%[g] (23.8%) |
14.7%[g] (19.6%) |
22.5%[g] (28.6%) |
28.7%[g] (37.2%) |
24.0%[g] (45.4%) |
35.1%[g] (58.1%) |
61.3%[g] (65.3%) |
61.2%[g] (68.0%) |
48.3%[g] (73.7%) |
| Belgium | Seq.[121][120] SGTF[121][120] |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
0.0% (<5%) |
0.0% (<5%) |
0.0% (<5%) |
13.6% (<5%) |
0.0% (<5%) |
7.1% (7.2%) |
7.7% (6.9%) |
13.4% (16.6%) |
23.3% (19.5%) |
39.4% (22.4%) |
43.2% (27.9%) |
51.5% (34.4%) |
57.6% (45.8%) |
66.5% (46.3%) |
79.3% (54.8%) |
| Portugal | SGTFL*Seq.%[75] | – | – | – | – | – | – | – | 1.7% | 0.8% | 1.2% | 1.7% | 2.9% | 6.8% | 12.3% | 22.7% | 33.5% | 39.2% | 40.9% | 45.7% | 50.5% | 56.0% | 64.3% |
| Sweden | Seq. all N501Y+A570D pos. tests[86] | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 10.8% (av. for 5 out of 21 regions)[h] |
15.1% (av. for 5 out of 21 regions)[i] |
27.3% (av. for 5 out of 21 regions)[j] |
30.4% (av. for 19 out of 21 regions)[k] |
41.5% (av. for 19 out of 21 regions)[l] |
56.4% (av. for 17 out of 21 regions)[m] |
71.3% (av. for 18 out of 21 regions)[n] |
| Germany | PCR proxy and Seq.[127] Seq.[127] |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (–) |
– (2.6%) |
2.0% (8.8%) |
3.6% (4.9%) |
4.7% (10.7%) |
7.2% (17.7%) |
17.6% (20.8%) |
25.9% (33.2%) |
40.0% (42.9%) |
54.5% (48.3%) |
63.5% (–) |
| Liechtenstein | PCR proxy: N501Y[134] | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 10.9% | 28.5% | 32.8% | 47.0% | 52.2% | 73.4% |
| Spain | Estimated based on PCR proxy[140] | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 5-10% (Jan.29)[135] |
– | – | 20-25% (Feb.18)[137] |
25-30% (Feb.22)[138] |
>50% (Mar.3)[139] |
– |
| United States | SGTF*Seq.%[175] | – | – | – | – | – | – | – | – | – | 0.05% | 0.2% | 0.4% | 0.5% | 1.0% | 1.9% | 3.0% | 4.7% | 7.6% | 12.4% | 19.3% | 28.8% | 37.4% |
| California | SGTF*Seq.%[175] | – | – | – | – | – | – | – | – | – | 0.3% | 0.3% | 0.7% | 1.1% | 1.3% | 2.0% | 1.9% | 4.2% | 5.9% | 13.1% | 16.6% | 18.8% | 28.0% |
| Florida | SGTF*Seq.%[175] | – | – | – | – | – | – | – | – | – | 0.2% | 0.5% | 0.9% | 1.0% | 2.3% | 4.6% | 8.1% | 10.7% | 14.5% | 21.0% | 28.3% | 39.8% | 47.0% |
| Other countries without weekly/periodic data, but with a variant share above 5%: Malta (61%),[131] and Poland (52%).[144] | |||||||||||||||||||||||
The following additional countries did not report variant shares, but are likely to have a significant share present due to their finding of more than 50 cases confirmed by whole genome sequencing per 13 March 2021:[176]
- Turkey (486 cases)
- Finland (268 cases)
- Australia (140 cases)
- Chile (124 cases)
- Croatia (121 cases)
- Nigeria (113 cases)
- South Korea (90 cases)
- Slovenia (87 cases)
- Latvia (83 cases)
- India (81 cases)
- Romania (76 cases)
- Ghana (67 cases)
- Singapore (66 cases)
- New Zealand (63 cases)
- North Macedonia (53 cases)
- Brazil (53 cases)
- Taiwan (32 cases)
The GISAID database of all sequenced COVID-19 genomes, calculates for each country for the past four weeks an average "Relative Variant Genome Frequency" for submitted samples. Those observed frequencies are however subject to sampling and reporting biases, and do not represent exact variant share prevalence due to absence of statistical representativity.[176]
Countries reporting a first case
December 2020
Cases of the variant began to be reported globally during December, being reported in Denmark,[64][177] Belgium,[178] the Netherlands, Australia[64][177] and Italy.[179] Shortly after, several other countries confirmed their first cases, the first of whom were found in Iceland and Gibraltar,[180][181] then Singapore, Israel and Northern Ireland on 23 December,[182][183][184] Germany and Switzerland on 24 December,[185][186] and the Republic of Ireland and Japan confirmed on 25 December.[187][188]
The first cases in Canada, France, Lebanon, Spain and Sweden were reported on 26 December.[189][190][191][192] Jordan, Norway, and Portugal reported their first case on 27 December,[193][194] Finland and South Korea reported their first cases on 28 December,[195][196] and Chile, India, Pakistan and the United Arab Emirates reported their first cases on 29 December.[197][198][199][200] The first case of new variant in Malta and Taiwan are reported on 30 December.[128][201] China and Brazil reported their first cases of the new variant on 31 December.[202][203] The United Kingdom and Denmark are sequencing their SARS-CoV-2 cases at considerably higher rates than most others,[204] and it was considered likely that additional countries would detect the variant later.[205]
The United States reported a case in Colorado with no travel history on 29 December, the sample was taken on 24 December.[206] On 6 January 2021, the US Centers for Disease Control and Prevention announced that it had found at least 52 confirmed cases in California, Florida, Colorado, Georgia, and New York.[207] In the following days, more cases of the variant were reported in other states, leading former CDC director Tom Frieden to express his concerns that the U.S. will soon face "close to a worst-case scenario".[208]
January 2021
Turkey detected its first cases in 15 people from England on 1 January 2021.[209] It was reported on 1 January that Denmark had found a total of 86 cases of the variant, equalling an overall frequency of less than 1% of the sequenced cases in the period from its first detection in the country in mid-November to the end of December;[210][211] this had increased to 1.6% of sequenced tests in the period from mid-November to week two of 2021, with 7% of sequenced tests in this week alone being of Alpha variant (lineage B.1.1.7).[212] Luxembourg and Vietnam reported their first case of this variant on 2 January 2021.[213][214]
On 3 January 2021, Greece and Jamaica detected their first four cases of this variant[215][216] and Cyprus announced that it had detected Alpha (VOC 202012/01) in 12 samples.[217] At the same time, New Zealand and Thailand reported their first cases of this variant, where the former reported six cases made up of five from the United Kingdom and one from South Africa,[218] and the latter reported the cases from a family of four who had arrived from Kent.[219] Georgia reported its first case[220] and Austria reported their first four cases of this variant, along with one case of Beta variant, on 4 January.[221]
On 5 January, Iran,[222] Oman,[223] and Slovakia reported their first cases of VOC-202012/01.[224] On 8 January, Romania reported its first case of the variant, an adult woman from Giurgiu County who declared not having left the country recently.[225] On 9 January, Peru confirmed its first case of the variant.[226] Mexico and Russia reported their first case of this variant on 10 January,[227] then Malaysia and Latvia on 11 January.[228][229]
On 12 January, Ecuador confirmed its first case of this variant.[230] The Philippines and Hungary both detected the presence of the variant on 13 January.[231][232] The Gambia recorded first cases of the variant on 14 January with it being the first confirmation of the variant's presence in Africa.[233] On 15 January, the Dominican Republic confirmed its first case of the new variant[234] and Argentina confirmed its first case of the variant on 16 January.[235] Czech Republic and Morocco reported their first cases on 18 January[236][237] while Ghana and Kuwait confirmed their first cases on 19 January.[238][239] Nigeria confirmed its first case on 25 January.[240] On 28 January, Senegal detected its first case of the variant.[241]
In early January, an outbreak linked to a primary school led to the detection of at least 30 cases of the new variant in the Bergschenhoek area of the Netherlands, signifying local transmission.[242]
On 24 January, a person travelling from Africa to Faroe Islands tested positive upon arrival to the islands, and went directly into quarantine.[243]
On 28 January, North Macedonia confirmed the variant was detected in a 46-year-old man, who had already recovered.[244]
February 2021
On 1 February, Lithuania confirmed the first cases of the new lineage.[citation needed] On 4 February, health authorities in Uruguay announced the first case of the variant in the country. The case was detected in a person who entered the country on 20 December 2020 and has been in quarantine ever since.[245] On 10 February, the Health Ministry of Croatia confirmed that out of 61 sequenced samples since 20 January, the variant was detected in 3 samples: a male 50 year old and 3.5 year old from Zagreb, and a male 34-year-old from Brod-Posavina County.[246] On 12 February, the variant was detected from four areas in Sri Lanka,[247] and the Canadian province of Newfoundland and Labrador confirmed an outbreak of the variant.[248]
March 2021
On 2 March, Indonesia reports its first cases of the variant in two migrant workers returning from Saudi Arabia.[249] On the same day, Tunisia[250] reported their first cases of the variant. The presence of the variant in Ivory Coast was confirmed on 25 March.[251]
N501Y mutation elsewhere
The N501Y mutation arose independently multiple times in different locations:
- In April 2020, it was seen for the first time in a few isolated sequences in Brazil.[252]
- In June 2020, the mutation appeared in an Australian lineage.[252]
- In July 2020, (according to Dr Julian Tang, University of Leicester) N501Y appeared in a lineage circulating in United States.[252][citation needed]
- In September 2020, it was found in a lineage in Wales, that evolved independently and was different from lineage B.1.1.7.[52]
- In September 2020,[1] the much more transmissible and widespread lineage B.1.1.7 (501Y.V1) was detected for the first time in Kent (England).[19]
- In October 2020, another highly transmissible variant of concern termed lineage B.1.351 (501Y.V2, Beta variant) was first time detected by genome sequencing in Nelson Mendela Bay (South Africa).[253] Phylogeographic analysis suggests this lineage emerged already in July or August 2020.[254]
- In December 2020,[255] another highly transmissible variant termed lineage P.1 (501Y.V3, Gamma variant) was detected in Manaus (Brazil).[256]
Statistics
Table
| Country | Confirmed cases (GISAID)[257] as of 16 January 2024 |
Last Case Reported |
|---|---|---|
| 268,555 | 27 June 2021 | |
| 250,995[258] | 6 July 2021 | |
| 219,985 | 29 June 2021 | |
| 102,642 | 28 June 2021 | |
| 62,570 | 29 June 2021 | |
| 58,838 | 21 June 2021 | |
| 32,782 | 28 June 2021 | |
| 28,773 | 25 June 2021 | |
| 32,899 | 1 July 2021 | |
| 25,420 | 30 June 2021 | |
| 21,726 | 26 June 2021 | |
| 22,258 | 5 July 2021 | |
| 20,724 | 1 July 2021 | |
| 14,804 | 21 June 2021 | |
| 15,798 | 23 June 2021 | |
| 9,334 | 11 June 2021 | |
| 7,988 | 24 June 2021 | |
| 8,526 | 23 June 2021 | |
| 8,366 | 20 June 2021 | |
| 7,953[259] | 25 May 2021 | |
| 5,481 | 7 June 2021 | |
| 5,015 | 23 June 2021 | |
| 4,480 | 19 June 2021 | |
| 4,898 | 25 May 2021 | |
| 4,350 | 11 June 2021 | |
| 3,836 | 20 June 2021 | |
| 4,299 | 24 June 2021 | |
| 3,361 | 1 June 2021 | |
| 3,135 | 28 May 2021 | |
| 2,993 | 8 May 2021 | |
| 3,056 | 17 May 2021 | |
| 1,705 | 24 June 2021 | |
| 726 | 20 June 2021 | |
| 767 | 15 June 2021 | |
| 1,001 | 30 March 2021 | |
| 637 | 25 May 2021 | |
| 735 | 8 June 2021 | |
| 551 | 16 June 2021 | |
| 584 | 1 June 2021 | |
| 506 | 4 July 2021 | |
| 559 | 3 June 2021 | |
| 470 | 26 June 2021 | |
| 384 | 24 June 2021 | |
| 361 | 12 June 2021 | |
| 318 | 19 May 2021 | |
| 386 | 25 June 2021 | |
| 347 | 24 April 2021 | |
| 258 | 22 April 2021 | |
| 231 | 23 May 2021 | |
| 190 | 17 June 2021 | |
| 179 | 28 May 2021 | |
| 183 | 15 June 2021 | |
| 162 | 27 April 2021 | |
| 152 | 14 June 2021 | |
| 148 | 11 June 2021 | |
| 162 | 12 June 2021 | |
| 140 | 19 March 2021 | |
| 165 | 17 June 2021 | |
| 131 | 8 March 2021 | |
| 139 | 7 May 2021 | |
| 167 | 17 June 2021 | |
| 159 | 7 June 2021 | |
| 110 | 10 March 2021 | |
| 106 | 22 April 2021 | |
| 129 | 24 May 2021 | |
| 93 | 8 June 2021 | |
| 99 | 25 May 2021 | |
| 90 | 4 May 2021 | |
| 68 | 21 June 2021 | |
| 48 | 9 March 2021 | |
| 87 | 8 June 2021 | |
| 80 | 8 June 2021 | |
| 79 | 23 April 2021 | |
| 72 | 25 March 2021 | |
| 63 | 28 June 2021 | |
| 70 | 18 March 2021 | |
| 52 | 8 May 2021 | |
| 54 | 14 June 2021 | |
| 59 | 22 May 2021 | |
| 39 | 11 May 2021 | |
| 44 | 5 May 2021 | |
| 31 | 5 February 2021 | |
| 59 | 29 May 2021 | |
| 62 | 31 March 2021 | |
| 29 | 26 February 2021 | |
| 28 | 11 March 2021 | |
| 22 | 9 April 2021 | |
| 25 | 6 May 2021 | |
| 66 | 6 April 2021 | |
| 45 | 11 March 2021 | |
| 21 | 5 June 2021 | |
| 154 | 25 May 2021 | |
| 21 | 25 February 2021 | |
| 19 | 29 December 2020 | |
| 26 | 21 May 2021 | |
| 20 | 5 January 2021 | |
| 28 | 29 April 2021 | |
| 35 | 30 April 2021 | |
| 17 | 15 June 2021 | |
| 767 | 3 May 2021 | |
| 15 | 9 March 2021 | |
| 152 | 12 February 2021 | |
| 15 | 14 May 2021 | |
| 7 | 18 March 2021 | |
| 11 | 17 April 2021 | |
| 7 | 17 May 2021 | |
| 25 | 30 March 2021 | |
| 31 | 4 June 2021 | |
| 11 | 2 June 2021 | |
| 10 | 27 January 2021 | |
| 10 | 19 April 2021 | |
| 9 | 4 May 2021 | |
| 16 | 28 March 2021 | |
| 16 | 14 April 2021 | |
| 17 | 7 April 2021 | |
| 14 | 16 May 2021 | |
| 7 | 21 January 2021 | |
| 6 | 21 May 2021 | |
| 12 | 6 May 2021 | |
| 10 | 15 May 2021 | |
| 5 | 22 March 2021 | |
| 16 | 1 April 2021 | |
| 4 | 8 April 2021 | |
| 4 | 11 June 2021 | |
| 3 | 7 February 2021 | |
| 3 | 24 February 2021 | |
| 3 | 3 February 2021 | |
| 3 | 4 February 2021 | |
| 22 | 6 January 2021 | |
| 148 | 10 June 2021 | |
| 16 | 3 March 2021 | |
| 3 | 17 April 2021 | |
| 29 | 5 March 2021 | |
| 6 | 8 March 2021 | |
| 2 | 23 March 2021 | |
| 2 | 5 January 2021 | |
| 2 | 17 March 2021 | |
| 2 | 4 May 2021 | |
| 3 | 8 April 2021 | |
| 2 | 8 March 2021 | |
| 2 | 21 May 2021 | |
| 2 | 5 February 2021 | |
| 2 | 28 May 2021 | |
| 6 | 5 March 2021 | |
| 2 | 11 February 2021 | |
| 1 | 20 January 2021 | |
| 1 | 19 April 2021 | |
| 1 | 9 February 2021 | |
| 17 | 7 April 2021 | |
| 1 | 19 May 2021 | |
| 1 | 23 April 2021 | |
| 1 | 10 January 2021 | |
| 1 | 21 April 2021 | |
| 1 | 29 April 2021 | |
| 30 | 22 December 2020 | |
| 4 | 15 April 2021 | |
| 1 | 9 January 2021 | |
| 1 | 5 June 2021 | |
| 1 | 3 April 2021 | |
| 1 | 27 May 2021 | |
| 1 | - | |
| 2 | 25 June 2021 | |
| 4 | 3 February 2021 | |
| 15 | 3 March 2021 | |
| 3 | March 2021 | |
| 4 | 29 March 2021 | |
| 3 | 7 May 2021 | |
| World (167 countries) | Total: 1,248,973 | Total as of 13 August 2021 |
Graphics
Confirmed cases by countries
- Note: The graphs presented here are only viewable by computers and some phones. If you cannot view it on your cell phone, switch to desktop mode from your browser.
- Data provided by various sources, such as; governmental, press or officials are updated every week since their last publication.
Graphs are unavailable due to technical issues. There is more info on Phabricator and on MediaWiki.org. |
Graphs are unavailable due to technical issues. There is more info on Phabricator and on MediaWiki.org. |
Graphs are unavailable due to technical issues. There is more info on Phabricator and on MediaWiki.org. |
Graphs are unavailable due to technical issues. There is more info on Phabricator and on MediaWiki.org. |
History
Detection

The Alpha variant (lineage B.1.1.7) was first detected in early December 2020 by analysing genome data with knowledge that the rates of infection in Kent were not falling despite national restrictions.[5][260]
The two earliest genomes that belong to lineage B.1.1.7 were collected on 20 September 2020 in Kent and another on 21 September 2020 in Greater London.[19] These sequences were submitted to the GISAID sequence database (sequence accessions EPI_ISL_601443 and EPI_ISL_581117, respectively).[261]
Backwards tracing using genetic evidence suggests lineage B.1.1.7 emerged in September 2020 and then circulated at very low levels in the population until mid-November. The increase in cases linked to the variant first became apparent in late November when Public Health England (PHE) was investigating why infection rates in Kent were not falling despite national restrictions. PHE then discovered a cluster linked to this variant spreading rapidly into London and Essex.[30]
Also important was the nature of the RT-PCR test used predominantly in the UK, Thermo Fisher Scientific's TaqPath COVID-19. The test matches RNA in three locations, and stopped working for the spike gene due to the HV 69–70 deletion—a deletion of the amino acids histidine and valine in positions 69 and 70, respectively[262]—in the spike protein of lineage B.1.1.7. This made preliminary identification easier because it could be better suspected which cases were with lineage B.1.1.7 through genome sequencing.[263]
It has been suggested that the variant may have originated in a chronically infected immunocompromised person, giving the virus a long time to replicate and evolve.[264][5][265][266]
Control
In the presence of a more transmissible variant, stronger physical distancing and lockdown measures were opted for to avoid overwhelming the population due to its tendency to grow exponentially.[267]
All countries of the United Kingdom were affected by domestic travel restrictions in reaction to the increased spread of the virus—at least partly attributed to Alpha—effective from 20 December 2020.[268][269] During December 2020, an increasing number of countries around the world either announced temporary bans on, or were considering banning, passenger travel from the UK, and in several cases from other countries such as the Netherlands and Denmark. Some countries banned flights; others allowed only their nationals to enter, subject to a negative SARS-CoV-2 test.[270] A WHO spokesperson said that, "[a]cross Europe, where transmission is intense and widespread, countries need to redouble their control and prevention approaches". Most bans by EU countries were for 48 hours, pending an integrated political crisis response meeting of EU representatives on 21 December to evaluate the threat from the new variant and coordinate a joint response.[271][272]
Many countries around the world imposed restrictions on passenger travel from the United Kingdom; neighbouring France also restricted manned goods vehicles (imposing a total ban before devising a testing protocol and permitting their passage once more).[273] Some also applied restrictions on travel from other countries.[274][275][276][277] As of 21 December 2020[update], at least 42 countries had restricted flights from the UK,[270] and Japan was restricting entry of all foreign nationals after cases of the new variant were detected in the country.[278]
The usefulness of travel bans has been contested as limited in cases where the variant has likely already arrived, especially if the estimated growth rate per week of the virus is higher locally.[279][280]
Extinction
In October 2021, Dr Jenny Harries, chief executive of the UK Health and Security Agency stated that previous circulating variants such as Alpha had 'disappeared' and replaced by the Delta variant.[281] In March 2022, the World Health Organization listed the Alpha, Beta and Gamma variants as previously circulating citing lack of any detected cases in the prior weeks and months.[282]
See also
Notes
- The difference between the two is explained by PHE:
SARS-CoV-2 variants, if considered to have concerning epidemiological, immunological, or pathogenic properties, are raised for formal investigation. At this point they are designated Variant Under Investigation (VUI) with a year, month, and number. Following a risk assessment with the relevant expert committee, they may be designated Variant of Concern (VOC).[18]
- SARS-CoV-2's S gene encodes its spike protein.[47]
- An example of this is the delta (Δ)–PCR test, which in connection to SARS-CoV-2, has been used to detect the HV 69–70 deletion in variants with this mutation[83] through what has been named "spike-gene target failure" (SGTF) or "spike-gene drop out"[75] for the spike (S) gene[c] in a subset of RT-PCR assays (e.g., TaqPath COVID-19 RT-PCR assay, ThermoFisher).[84] Though existing in a few other variants of SARS-CoV-2,[75] the HV 69–70 deletion in the spike protein is present in the vast majority of B.1.1.7 genomes, which enables the delta-PCR test to be used as a proxy test for the lineage—or as a supplementing first-screening test before conducting the whole-genome sequencing.[83][75]
Another example of a RT-PCR test intended to detect specific variants is the one detecting all genomes with the N501Y mutation (e.g., Alpha, Gamma variant, and Beta variant), which is now also being used as a first-step screening tool ahead of genome sequencing by several laboratories/countries (e.g. by some parts of France).[85]
A third and fourth RT-PCR test intended for detecting specific variants pre-screen the samples for variants respectively with the N501Y+A570D mutations (Alpha) and N501Y without the A570D mutation (Beta, Gamma, and other N501Y variants).[86] - 104 WHO members had reported a detection of the Alpha variant (lineage B.1.1.7), as of 23 March 2021: Albania, Angola, Argentina, Australia, Austria, Azerbaijan, Bahrain, Bangladesh, Barbados, Belarus, Belgium, Belize, Bosnia and Herzegovina, Brazil, Brunei Darussalam, Bulgaria, Cabo Verde, Cambodia, Canada, Chile, China, Costa Rica, Croatia, Cyprus, Czech Republic, Democratic Republic of the Congo, Denmark, Dominican Republic, Ecuador, Estonia, Finland, France, Gambia, Georgia, Germany, Ghana, Greece, Hungary, Iceland, India, Indonesia, Iran, Iraq, Ireland, Israel, Italy, Jamaica, Japan, Jordan, Kenya, Kuwait, Latvia, Lebanon, Libya, Lithuania, Luxembourg, Malaysia, Malta, Mauritania, Mauritius, Mexico, Monaco, Montenegro, Morocco, Nepal, Netherlands, New Zealand, Nigeria, North Macedonia, Norway, Oman, Pakistan, Peru, Philippines, Poland, Portugal, Republic of Korea (South Korea), Republic of Moldova, Romania, Russia, Rwanda, Saint Lucia, Saudi Arabia, Senegal, Serbia, Singapore, Slovakia, Slovenia, South Africa, Spain, Sri Lanka, Sweden, Switzerland, Thailand, Trinidad and Tobago, Tunisia, Turkey, Ukraine, United Arab Emirates, United Kingdom, United States, Uruguay, Uzbekistan, Vietnam.[87]
90 WHO members had not reported any detection of the Alpha variant (lineage B.1.1.7), as of 23 March 2021: Afghanistan, Algeria, Andorra, Antigua and Barbuda, Armenia, Bahamas, Benin, Bhutan, Bolivia, Botswana, Burkina Faso, Burundi, Cameroon, Central African Republic, Chad, Colombia, Comoros, Congo, Cook Islands, Côte d'Ivoire, Cuba, Democratic People's Republic of Korea (North Korea), Djibouti, Dominica, Egypt, El Salvador, Equatorial Guinea, Eritrea, Ethiopia, Fiji, Gabon, Grenada, Guatemala, Guinea-Bissau, Guyana, Haiti, Honduras, Guinea, Kazakhstan, Kiribati, Kyrgyzstan, Lao People's Democratic Republic, Lesotho, Liberia, Madagascar, Malawi, Maldives, Mali, Marshall Islands, Micronesia, Mongolia, Mozambique, Myanmar, Namibia, Nauru, Nicaragua, Niger, Niue, Palau, Panama, Papua New Guinea, Paraguay, Qatar, Saint Kitts and Nevis, Saint Vincent and the Grenadines, Samoa, San Marino, Sao Tome and Principe, Seychelles, Sierra Leone, Solomon Islands, Somalia, South Sudan, Sudan, Suriname, Swaziland, Syrian Arab Republic, Tajikistan, Timor-Leste, Togo, Tonga, Turkmenistan, Tuvalu, Uganda, United Republic of Tanzania, Vanuatu, Venezuela, Yemen, Zambia, Zimbabwe.[87] - The weekly UK infection survey lists for each country in Great Britain a set of raw data and average-smoothed modelled data from SGTF analysed PCR tests collected from private households (excl. tests from hospitals, care centres and public institutions). Raw data as well as model data for Wales, Northern Ireland and Scotland must be treated with caution due to a small number of collected samples, resulting in great data uncertainty.[165][166]
- All PCR tests were analysed for 3 genes present in the coronavirus: N protein, S protein and ORF1ab (see Table 6A in the Infection Survey). Each PCR test can have any one, any two or all three genes detected. Coronavirus positives are those where one or more of these genes is detected in the swab (other than tests that are only positive on the S-gene which is not considered a reliable indicator of the virus if found on its own). The new B.1.1.7 variant of COVID-19 has genetic changes in the S gene, that results in the S-gene no longer being detected in the current test, meaning that it will only be positive on the ORF1ab and the N gene. The survey uses the terms "New UK variant compatible" for ORF1ab + N protein gene positives, "not compatible with new UK variant" for ORF1ab + N protein + S protein gene positive, and "virus too low to be identifiable" for all other gene patterns (a reasonable definition given that all samples taken from the first phase of the COVID-19 disease where virus exist by identifiable quantity, either will be positive by "ORF1ab+N" or "ORF1ab+N+S"). However, further uncertainty exists given that not all "New UK variant compatible" SGTF cases (positive on ORF1ab and N-genes, but not the S-gene) will be the new UK variant - due to some other competing variants also delivering this same test pattern; and prior to mid-November 2020 the data should not be read as being an indicator of the variant at all.[165][166]
- The weekly B.1.1.7 variant share raw data is calculated from Table 6A, by dividing the "ORF1ab+N" percentage with the percentage sum of "ORF1ab+N" and "ORF1ab+N+S". Table 6C utilized the raw data from table 6A as input for calculation of some modelled (average-smoothed and weighted) daily estimated figures for the respective percentage of the population being coronavirus positive by either the "New UK variant compatible virus", or a "Not compatible with new UK variant virus" or a "virus too low to be identifiable", with data from earlier dates in 2020 also being available when downloading the published earlier editions of the Infection Survey. The modelled weekly average value for the Alpha variant, noted in this table as the value (m: %), is calculated as the average for the listed seven days in each week of "New UK variant compatible percentage" divided by the sum of "New UK variant compatible percentage" and "Not compatible with new UK variant percentage".[165][166]
- In Austria, the listed B.1.1.7 variant shares represent the percentage of all positive COVID-19 tests being confirmed to be B.1.1.7 by genome sequencing. However, many N501Y positive RT-PCR tests were not variant determined further by genome sequencing, and likely would have returned a B.1.1.7 positive result for the vast majority of such tests if the genome sequencing analysis had been done. If all N501Y positive tests had been analysed further by genome sequencing, then the listed weekly B.1.1.7 shares could have been uptil: 9.6% for week 1 (2.4% higher), 23.8% for week 2 (6.4% higher), 19.6% for week 3 (4.9% higher), 28.6% for week 4 (6.1% higher), 37.2% for week 5 (8.5% higher), 45.4% for week 6 (21.4% higher), 58.1% for week 7 (23.0% higher), 65.3% for week 8 (4.0% higher), 68.0% for week 9 (6.8% higher) and 73.7% for week 10 (25.4% higher).[126]
- In Sweden, a study comprising 11% of all SARS-CoV-2–positive PCR samples nationwide for week 4 found the B.1.1.7 share to be 10.8% (243/2244). Samples were, however, only collected from five southern regions (Skåne, Västra Götaland, Västmanland, Gävleborg and Örebro), which were not considered to be statistically representative for the demography and geography of Sweden as a whole. The national authorities plan to expand the weekly study to cover more regions for the following weeks in February 2021.[174][133]
- In Sweden, a study comprising 16% of all SARS-CoV-2–positive PCR samples nationwide for week 5 found the B.1.1.7 share to be 15.1% (488/3224). Samples were, however, only collected from five southern regions (Skåne, Västra Götaland, Västmanland, Gävleborg and Örebro), which were not considered to be statistically representative for the demography and geography of Sweden as a whole. The national authorities also calculated a two-week average (week 5+6) for 19 out of 21 regions, and plan to expand the weekly study to cover more regions for the following weeks in February 2021.[133]
- In Sweden, a study comprising 18% of all SARS-CoV-2–positive PCR samples nationwide for week 6 found the B.1.1.7 share to be 27.3% (1021/3742). Samples were, however, only collected from five southern regions (Skåne, Västra Götaland, Västmanland, Gävleborg and Örebro), which were not considered to be statistically representative for the demography and geography of Sweden as a whole. The national authorities also calculated a two-week average (week 5+6) for 19 out of 21 regions, and plan to expand the weekly study to cover more regions for the following weeks in February 2021.[133]
- In Sweden, a study comprising 47.8% of all SARS-CoV-2–positive PCR samples nationwide for week 7, collected samples from 19 out of 21 regions (all except Gotland and Västerbotten), and found the B.1.1.7 share to be 30.4% (3316/10910) as a simple overall average (with a potential geographically skewed misrepresentation in the calculation as no data weights were used to normalize/correct each regions share of the overall sample).[86]
- In Sweden, a study comprising 42.2% of all SARS-CoV-2–positive PCR samples nationwide for week 8, collected samples from 19 out of 21 regions (all except Gotland and Västerbotten), and found the B.1.1.7 share to be 41.5% (4643/11191) as a simple overall average (with a potential geographically skewed misrepresentation in the calculation as no data weights were used to normalize/correct each regions share of the overall sample).[86]
- In Sweden, a study comprising 37.8% of all SARS-CoV-2–positive PCR samples nationwide for week 9, collected samples from 17 out of 21 regions (all except Gotland, Västerbotten, Norrbotten and Östergötland), and found the B.1.1.7 share to be 56.4% (5939/10528) as a simple overall average (with a potential geographically skewed misrepresentation in the calculation as no data weights were used to normalize/correct each regions share of the overall sample).[86]
- In Sweden, a study comprising 43.1% of all SARS-CoV-2–positive PCR samples nationwide for week 10, collected samples from 18 out of 21 regions (all except Gotland, Västerbotten and Östergötland), and found the B.1.1.7 share to be 71.3% (8850/12417) as a simple overall average (with a potential geographically skewed misrepresentation in the calculation as no data weights were used to normalize/correct each regions share of the overall sample).[86]
- "UK reports new variant, termed VUI 202012/01". GISAID. Archived from the original on 20 December 2020. Retrieved 20 December 2020.
- Threat Assessment Brief: Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom (PDF) (Report). European Centre for Disease Prevention and Control (ECDC). 20 December 2020.
References
External links
Wikiwand in your browser!
Seamless Wikipedia browsing. On steroids.
Every time you click a link to Wikipedia, Wiktionary or Wikiquote in your browser's search results, it will show the modern Wikiwand interface.
Wikiwand extension is a five stars, simple, with minimum permission required to keep your browsing private, safe and transparent.