Loading AI tools
Demethylases are enzymes that remove methyl (CH3) groups from nucleic acids, proteins (particularly histones), and other molecules. Demethylases are important epigenetic proteins, as they are responsible for transcriptional regulation of the genome by controlling the methylation of DNA and histones, and by extension, the chromatin state at specific gene loci.

Histone methylation was initially considered an effectively irreversible process as the half-life of the histone methylation was approximately equal to the histone half-life.[1] Histone lysine demethylase LSD1 (later classified as KDM1A) was first identified in 2004 as a nuclear amine oxidase homolog.[2] Two main classes of histone lysine demethylases exist, defined by their mechanisms: flavin adenine dinucleotide (FAD)-dependent amine oxidases and α-ketoglutarate-dependent hydroxylases.
Histone lysine demethylases possess a variety of domains that are responsible for histone recognition, DNA binding, methylated amino acid substrate binding and catalytic activity. These include:
- FAD-dependent amine oxidase domains containing the active catalytic site of KDM1
- Jumonji-C domains containing the active catalytic site of KDM2 through KDM8[3][4]
- Jumonji-N domains responsible for Jumonji-C domain conformation stability
- SWIRM (SWI3P, RSC8P and Moira) domains proposed as an anchor site for histone substrates and responsible for chromatin stability
- PHD, CXXC and C5HC2 zinc finger domains responsible for histone recognition and binding
Histone lysine demethylases are classified according to their domains and unique substrate specificities. The lysine substrates and identified according to their position in the corresponding histone amino acid sequence and methylation state (for example, H3K9me3 refers to trimethylated histone 3 lysine 9.)


- KDM1
- The KDM1 homologs include KDM1A and KDM1B. KDM1A demethylates H3K4me1/2 and H3K9me1/2, and KDM1B emethylates H3K4me1/2. KDM1 activity is critical to embryogenesis and tissue-specific differentiation, as well as oocyte growth.[1] Deletion of the gene for KDM1A can have effects on the growth and differentiation of embryonic stem cells and is universally lethal in knockout mice.[5][6] KDM1A gene expression is observed to be upregulated in some cancers,[7][8] and so KDM1A inhibition has therefore been considered a possible epigenetic treatment for cancer.[9][10][11]:KDM1B, however, is mostly involved in oocyte development. Deletion of this gene leads to maternal effect lethality in mice.[12] Orthologs of KDM1 in D. melanogaster and C. elegans appear to function similarly to KDM1B rather than KDM1A.[13][14]
- KDM2
- The KDM2 homologs include KDM2A and KDM2B. KDM2A and KDM2B demethylate H3K4me3 and H3K36me2/3. KDM2A has roles in either promoting or inhibiting tumor function, and KDM2B has roles in oncogenesis.[1] KDM2A and KDM2B possess CXXC zinc finger domains responsible for binding to unmethylated CpG islands, and it is believed that they may bind to many gene regulatory elements in the absence of sequence-specific transcription factors.[15]:Overexpressed KDM2B has been observed in human lymphoma and adenocarcinoma, and underexpressed KDM2B has been observed in human prostate cancer and glioblastoma. KDM2B has been additionally shown to prevent senescence in some cells through ectopic expression.[16]
- KDM3
- The KDM3 homologs include KDM3A, KDM3B and KDM3C. KDM3A, KDM3B and KDM3C demethylate H3K9me1/2. KDM3A has roles in spermatogenesis and metabolic functions, however, the activity of KDM3B and KDM3C are not specifically known.[1]:Knockdown studies of KDM3A in mice resulted in male infertility and adult onset-obesity. Additional studies have indicated that KDM3A may play a role in regulation of androgen receptor-dependent genes as well as genes involved in pluripotency, indicating a potential role for KDM3A in tumorigenesis.[17]
- KDM4
- The KDM4 homologs include KDM4A, KDM4B, KDM4C, KDM4D, KDM4E and KDM4F. KDM4A, KDM4B and KDM4C demethylate H3K9me2/3, H3K9me3 and H3K36me2/3, and KDM4D, KDM4E and KDM4F demethylate H3K9me2/3. KDM4A, KDM4B, KDM4C and KDM4D have roles in tumorigenesis, however, the activity of KDM4E and KDM4F are not specifically known..[1] KDM4B upregulation has bee observed in medulloblastoma, and KDM4C amplification has been documented in oesophageal squamous carcinoma, medulloblastoma and breast cancer.[18][19][20][21] Other gene expression data has also suggested KDM4A, KDM4B, and KDM4C are overexpressed in prostate cancer.[22]
- KDM5
- The KDM5 homologs includes KDM5A, KDM5B, KDM5C and KDM5D. KDM5A, KDM5B, KDM5C and KDM5D demethylate H3K4me2/3.[1] The KDM5 family appears to regulate key developmental functions, including cellular differentiation, mitochondrial function and cell cycle progression.[23][24][25][26][27][28] KDM5B and KDM5C have also shown to interaction with PcG proteins, which are involved in transcriptional repression. KDM5C mutations on the X-chromosome have also been observed in patients with X-linked intellectual disability.[29] Depletion of KDM5C homologs in D. rerio have shown brain-patterning defects and neuronal cell death.[30]
- KDM6
- The KDM6 family includes KDM6A, KDM6B and KDM6C. KDM6A and KDM6B demethylate H3K27me2/3, and KDM4C demethylates H3K27me3. KDM6A and KDM6B possess tumor-suppressive characteristics. KDM6A knockdowns in fibroblasts lead to an immediate increase in fibroblast population. KDM6B expressed in fibroblasts induces oncogenes of the Ras/Raf/MEK/ERK pathway.[31] Point mutations of KDM6A have been identified as one cause of Kabuki syndrome, a congenital disorder resulting in intellectual disability.[32][33] Deletion of KDM6A in D. rerio results in decreased expression of HOX genes, which play a role in regulating body patterning during development.[34] In mammalian studies, KDM6A has been shown to regulate HOX genes as well.[35][36] Mutation of KDM5B disrupt gonad development in C.elegans.[35] Other studies have shown that KDM6B expression is upregulated in activated macrophages and dynamically expressed during differentiation of stem cells.[37][38]

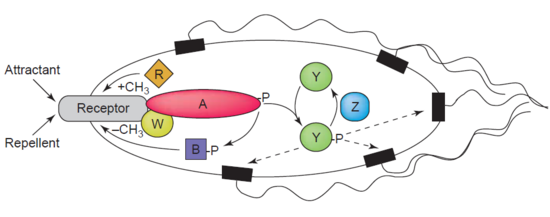
Another example of a demethylase is protein-glutamate methylesterase, also known as CheB protein (EC 3.1.1.61), which demethylates MCPs (methyl-accepting chemotaxis proteins) through hydrolysis of carboxylic ester bonds. The association of a chemotaxis receptor with an agonist leads to the phosphorylation of CheB. Phosphorylation of CheB protein enhances its catalytic MCP demethylating activity resulting in adaption of the cell to environmental stimuli.[39] MCPs respond to extracellular attractants and repellents in bacteria like E. coli in chemotaxis regulation. CheB is more specifically termed a methylesterase, as it removes methyl groups from methylglutamate residues located on the MCPs through hydrolysis, producing glutamate accompanied by the release of methanol.[40]
CheB is of particular interest to researchers as it may be a therapeutic target for mitigating the spread of bacterial infections.[41]
Wikiwand in your browser!
Seamless Wikipedia browsing. On steroids.
Every time you click a link to Wikipedia, Wiktionary or Wikiquote in your browser's search results, it will show the modern Wikiwand interface.
Wikiwand extension is a five stars, simple, with minimum permission required to keep your browsing private, safe and transparent.