Top Qs
Timeline
Chat
Perspective
Notch signaling pathway
Series of molecular signals From Wikipedia, the free encyclopedia
Remove ads
The Notch signaling pathway is a highly conserved cell signaling system present in most animals.[1] Mammals possess four different notch receptors, referred to as NOTCH1, NOTCH2, NOTCH3, and NOTCH4.[2] The notch receptor is a single-pass transmembrane receptor protein. It is a hetero-oligomer composed of a large extracellular portion, which associates in a calcium-dependent, non-covalent interaction with a smaller piece of the notch protein composed of a short extracellular region, a single transmembrane-pass, and a small intracellular region.[3]
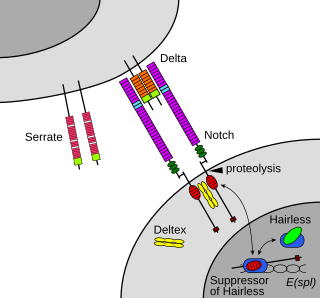

Notch signaling promotes proliferative signaling during neurogenesis, and its activity is inhibited by Numb to promote neural differentiation. It plays a major role in the regulation of embryonic development.
Notch signaling is dysregulated in many cancers, and faulty notch signaling is implicated in many diseases, including T-cell acute lymphoblastic leukemia (T-ALL),[4] cerebral autosomal-dominant arteriopathy with sub-cortical infarcts and leukoencephalopathy (CADASIL), multiple sclerosis, Tetralogy of Fallot, and Alagille syndrome. Inhibition of notch signaling inhibits the proliferation of T-cell acute lymphoblastic leukemia in both cultured cells and a mouse model.[5][6]
Remove ads
Discovery
In 1914, John S. Dexter noticed the appearance of a notch in the wings of the fruit fly Drosophila melanogaster. The alleles of the gene were identified in 1917 by American evolutionary biologist Thomas Hunt Morgan.[7][8] Its molecular analysis and sequencing was independently undertaken in the 1980s by Spyros Artavanis-Tsakonas and Michael W. Young.[9][10] Alleles of the two C. elegans Notch genes were identified based on developmental phenotypes: lin-12[11] and glp-1.[12][13] The cloning and partial sequence of lin-12 was reported at the same time as Drosophila Notch by Iva Greenwald.[14]
Remove ads
Mechanism
Summarize
Perspective
The Notch protein spans the cell membrane, with part of it inside and part outside. Ligand proteins binding to the extracellular domain induce proteolytic cleavage and release of the intracellular domain, which enters the cell nucleus to modify gene expression.[15]
The cleavage model was first proposed in 1993 based on work done with Drosophila Notch and C. elegans lin-12,[16][17] informed by the first oncogenic mutation affecting a human Notch gene.[18] Compelling evidence for this model was provided in 1998 by in vivo analysis in Drosophila by Gary Struhl[19] and in cell culture by Raphael Kopan.[20] Although this model was initially disputed,[1] the evidence in favor of the model was irrefutable by 2001.[21][22]
The receptor is normally triggered via direct cell-to-cell contact, in which the transmembrane proteins of the cells in direct contact form the ligands that bind the notch receptor. The Notch binding allows groups of cells to organize themselves such that, if one cell expresses a given trait, this may be switched off in neighbouring cells by the intercellular notch signal. In this way, groups of cells influence one another to make large structures. Thus, lateral inhibition mechanisms are key to Notch signaling. lin-12 and Notch mediate binary cell fate decisions, and lateral inhibition involves feedback mechanisms to amplify initial differences.[21]
The Notch cascade consists of Notch and Notch ligands, as well as intracellular proteins transmitting the notch signal to the cell's nucleus. The Notch/Lin-12/Glp-1 receptor family[23] was found to be involved in the specification of cell fates during development in Drosophila and C. elegans.[24]
The intracellular domain of Notch forms a complex with CBF1 and Mastermind to activate transcription of target genes. The structure of the complex has been determined.[25][26]
Pathway
Maturation of the notch receptor involves cleavage at the prospective extracellular side during intracellular trafficking in the Golgi complex.[27] This results in a bipartite protein, composed of a large extracellular domain linked to the smaller transmembrane and intracellular domain. Binding of ligand promotes two proteolytic processing events; as a result of proteolysis, the intracellular domain is liberated and can enter the nucleus to engage other DNA-binding proteins and regulate gene expression.
Notch and most of its ligands are transmembrane proteins, so the cells expressing the ligands typically must be adjacent to the notch expressing cell for signaling to occur.[citation needed] The notch ligands are also single-pass transmembrane proteins and are members of the DSL (Delta/Serrate/LAG-2) family of proteins. In Drosophila melanogaster (the fruit fly), there are two ligands named Delta and Serrate. In mammals, the corresponding names are Delta-like and Jagged. In mammals there are multiple Delta-like and Jagged ligands, as well as possibly a variety of other ligands, such as F3/contactin.[28]
In the nematode C. elegans, two genes encode homologous proteins, glp-1 and lin-12. There has been at least one report that suggests that some cells can send out processes that allow signaling to occur between cells that are as much as four or five cell diameters apart.[citation needed]
The notch extracellular domain is composed primarily of small cystine-rich motifs called EGF-like repeats.[29]
Notch 1, for example, has 36 of these repeats. Each EGF-like repeat is composed of approximately 40 amino acids, and its structure is defined largely by six conserved cysteine residues that form three conserved disulfide bonds. Each EGF-like repeat can be modified by O-linked glycans at specific sites.[30] An O-glucose sugar may be added between the first and second conserved cysteines, and an O-fucose may be added between the second and third conserved cysteines. These sugars are added by an as-yet-unidentified O-glucosyltransferase (except for Rumi), and GDP-fucose Protein O-fucosyltransferase 1 (POFUT1), respectively. The addition of O-fucose by POFUT1 is absolutely necessary for notch function, and, without the enzyme to add O-fucose, all notch proteins fail to function properly. As yet, the manner by which the glycosylation of notch affects function is not completely understood.
The O-glucose on notch can be further elongated to a trisaccharide with the addition of two xylose sugars by xylosyltransferases, and the O-fucose can be elongated to a tetrasaccharide by the ordered addition of an N-acetylglucosamine (GlcNAc) sugar by an N-Acetylglucosaminyltransferase called Fringe, the addition of a galactose by a galactosyltransferase, and the addition of a sialic acid by a sialyltransferase.[31]
To add another level of complexity, in mammals there are three Fringe GlcNAc-transferases, named lunatic fringe, manic fringe, and radical fringe. These enzymes are responsible for something called a "fringe effect" on notch signaling.[32] If Fringe adds a GlcNAc to the O-fucose sugar then the subsequent addition of a galactose and sialic acid will occur. In the presence of this tetrasaccharide, notch signals strongly when it interacts with the Delta ligand, but has markedly inhibited signaling when interacting with the Jagged ligand.[33] The means by which this addition of sugar inhibits signaling through one ligand, and potentiates signaling through another is not clearly understood.
Once the notch extracellular domain interacts with a ligand, an ADAM-family metalloprotease called ADAM10, cleaves the notch protein just outside the membrane.[34] This releases the extracellular portion of notch (NECD), which continues to interact with the ligand. The ligand plus the notch extracellular domain is then endocytosed by the ligand-expressing cell. There may be signaling effects in the ligand-expressing cell after endocytosis; this part of notch signaling is a topic of active research.[citation needed] After this first cleavage, an enzyme called γ-secretase (which is implicated in Alzheimer's disease) cleaves the remaining part of the notch protein just inside the inner leaflet of the cell membrane of the notch-expressing cell. This releases the intracellular domain of the notch protein (NICD), which then moves to the nucleus, where it can regulate gene expression by activating the transcription factor CSL. It was originally thought that these CSL proteins suppressed Notch target transcription. However, further research showed that, when the intracellular domain binds to the complex, it switches from a repressor to an activator of transcription.[35] Other proteins also participate in the intracellular portion of the notch signaling cascade.[36]
Ligand interactions

Notch signaling is initiated when Notch receptors on the cell surface engage ligands presented in trans on opposing cells. Despite the expansive size of the Notch extracellular domain, it has been demonstrated that EGF domains 11 and 12 are the critical determinants for interactions with Delta.[37] Additional studies have implicated regions outside of Notch EGF11-12 in ligand binding. For example, Notch EGF domain 8 plays a role in selective recognition of Serrate/Jagged[38] and EGF domains 6-15 are required for maximal signaling upon ligand stimulation.[39] A crystal structure of the interacting regions of Notch1 and Delta-like 4 (Dll4) provided a molecular-level visualization of Notch-ligand interactions, and revealed that the N-terminal MNNL (or C2) and DSL domains of ligands bind to Notch EGF domains 12 and 11, respectively.[40] The Notch1-Dll4 structure also illuminated a direct role for Notch O-linked fucose and glucose moieties in ligand recognition, and rationalized a structural mechanism for the glycan-mediated tuning of Notch signaling.[40]
Synthetic Notch signaling
It is possible to engineer synthetic Notch receptors by replacing the extracellular receptor and intracellular transcriptional domains with other domains of choice. This allows researchers to select which ligands are detected, and which genes are upregulated in response. Using this technology, cells can report or change their behavior in response to contact with user-specified signals, facilitating new avenues of both basic and applied research into cell-cell signaling.[41] Notably, this system allows multiple synthetic pathways to be engineered into a cell in parallel.[42][43]
Remove ads
Function
Summarize
Perspective
The Notch signaling pathway is important for cell-cell communication, which involves gene regulation mechanisms that control multiple cell differentiation processes during embryonic and adult life. Notch signaling also has a role in the following processes:
- neuronal function and development[44][45][46][47]
- stabilization of arterial endothelial fate and angiogenesis[48]
- regulation of crucial cell communication events between endocardium and myocardium during both the formation of the valve primordial and ventricular development and differentiation[49]
- cardiac valve homeostasis, as well as implications in other human disorders involving the cardiovascular system[50]
- timely cell lineage specification of both endocrine and exocrine pancreas[51]
- influencing of binary fate decisions of cells that must choose between the secretory and absorptive lineages in the gut[52]
- expansion of the hematopoietic stem cell compartment during bone development and participation in commitment to the osteoblastic lineage, suggesting a potential therapeutic role for notch in bone regeneration and osteoporosis[53]
- expansion of the hemogenic endothelial cells along with signaling axis involving Hedgehog signaling and Scl[54]
- T cell lineage commitment from common lymphoid precursor [55]
- regulation of cell-fate decision in mammary glands at several distinct development stages[56]
- possibly some non-nuclear mechanisms, such as control of the actin cytoskeleton through the tyrosine kinase Abl[28]
- Regulation of the mitotic/meiotic decision in the C. elegans germline[12]
- development of alveoli in the lung.[57]
It has also been found that Rex1 has inhibitory effects on the expression of notch in mesenchymal stem cells, preventing differentiation.[58]
Role in embryogenesis
Summarize
Perspective
The Notch signaling pathway plays an important role in cell-cell communication, and further regulates embryonic development.
Embryo polarity
Notch signaling is required in the regulation of polarity. For example, mutation experiments have shown that loss of Notch signaling causes abnormal anterior-posterior polarity in somites.[59] Also, Notch signaling is required during left-right asymmetry determination in vertebrates.[60]
Early studies in the nematode model organism C. elegans indicate that Notch signaling has a major role in the induction of mesoderm and cell fate determination.[12] As mentioned previously, C. elegans has two genes that encode for partially functionally redundant Notch homologs, glp-1 and lin-12.[61] During C. elegans, GLP-1, the C. elegans Notch homolog, interacts with APX-1, the C. elegans Delta homolog. This signaling between particular blastomeres induces differentiation of cell fates and establishes the dorsal-ventral axis.[62]
Role in somitogenesis
Notch signaling is central to somitogenesis. In 1995, Notch1 was shown to be important for coordinating the segmentation of somites in mice.[63] Further studies identified the role of Notch signaling in the segmentation clock. These studies hypothesized that the primary function of Notch signaling does not act on an individual cell, but coordinates cell clocks and keep them synchronized. This hypothesis explained the role of Notch signaling in the development of segmentation and has been supported by experiments in mice and zebrafish.[64][65][66] Experiments with Delta1 mutant mice that show abnormal somitogenesis with loss of anterior/posterior polarity suggest that Notch signaling is also necessary for the maintenance of somite borders.[63]
During somitogenesis, a molecular oscillator in paraxial mesoderm cells dictates the precise rate of somite formation. A clock and wavefront model has been proposed in order to spatially determine the location and boundaries between somites. This process is highly regulated as somites must have the correct size and spacing in order to avoid malformations within the axial skeleton that may potentially lead to spondylocostal dysostosis. Several key components of the Notch signaling pathway help coordinate key steps in this process. In mice, mutations in Notch1, Dll1 or Dll3, Lfng, or Hes7 result in abnormal somite formation. Similarly, in humans, the following mutations have been seen to lead to development of spondylocostal dysostosis: DLL3, LFNG, or HES7.[67]
Role in epidermal differentiation
Notch signaling is known to occur inside ciliated, differentiating cells found in the first epidermal layers during early skin development.[68] Furthermore, it has found that presenilin-2 works in conjunction with ARF4 to regulate Notch signaling during this development.[69] However, it remains to be determined whether gamma-secretase has a direct or indirect role in modulating Notch signaling.
Remove ads
Role in central nervous system development and function
Summarize
Perspective

Early findings on Notch signaling in central nervous system (CNS) development were performed mainly in Drosophila with mutagenesis experiments. For example, the finding that an embryonic lethal phenotype in Drosophila was associated with Notch dysfunction[70] indicated that Notch mutations can lead to the failure of neural and Epidermal cell segregation in early Drosophila embryos. In the past decade, advances in mutation and knockout techniques allowed research on the Notch signaling pathway in mammalian models, especially rodents.
The Notch signaling pathway was found to be critical mainly for neural progenitor cell (NPC) maintenance and self-renewal. In recent years, other functions of the Notch pathway have also been found, including glial cell specification,[71][72] neurites development,[73] as well as learning and memory.[74]
Neuron cell differentiation
The Notch pathway is essential for maintaining NPCs in the developing brain. Activation of the pathway is sufficient to maintain NPCs in a proliferating state, whereas loss-of-function mutations in the critical components of the pathway cause precocious neuronal differentiation and NPC depletion.[45] Modulators of the Notch signal, e.g., the Numb protein are able to antagonize Notch effects, resulting in the halting of cell cycle and the differentiation of NPCs.[75][76] Conversely, the fibroblast growth factor pathway promotes Notch signaling to keep stem cells of the cerebral cortex in the proliferative state, amounting to a mechanism regulating cortical surface area growth and, potentially, gyrification.[77][78] In this way, Notch signaling controls NPC self-renewal as well as cell fate specification.
A non-canonical branch of the Notch signaling pathway that involves the phosphorylation of STAT3 on the serine residue at amino acid position 727 and subsequent Hes3 expression increase (STAT3-Ser/Hes3 Signaling Axis) has been shown to regulate the number of NPCs in culture and in the adult rodent brain.[79]
In adult rodents and in cell culture, Notch3 promotes neuronal differentiation, having a role opposite to Notch1/2.[80] This indicates that individual Notch receptors can have divergent functions, depending on cellular context.
Neurite development
In vitro studies show that Notch can influence neurite development.[73] In vivo, deletion of the Notch signaling modulator, Numb, disrupts neuronal maturation in the developing cerebellum,[81] whereas deletion of Numb disrupts axonal arborization in sensory ganglia.[82] Although the mechanism underlying this phenomenon is not clear, together these findings suggest Notch signaling might be crucial in neuronal maturation.
Gliogenesis
In gliogenesis, Notch appears to have an instructive role that can directly promote the differentiation of many glial cell subtypes.[71][72] For example, activation of Notch signaling in the retina favors the generation of Muller glia cells at the expense of neurons, whereas reduced Notch signaling induces production of ganglion cells, causing a reduction in the number of Muller glia.[45]
Adult brain function
Apart from its role in development, evidence shows that Notch signaling is also involved in neuronal apoptosis, neurite retraction, and neurodegeneration of ischemic stroke in the brain[83] In addition to developmental functions, Notch proteins and ligands are expressed in cells of the adult nervous system,[84] suggesting a role in CNS plasticity throughout life. Adult mice heterozygous for mutations in either Notch1 or Cbf1 have deficits in spatial learning and memory.[74] Similar results are seen in experiments with presenilins1 and 2, which mediate the Notch intramembranous cleavage. To be specific, conditional deletion of presenilins at 3 weeks after birth in excitatory neurons causes learning and memory deficits, neuronal dysfunction, and gradual neurodegeneration.[85] Several gamma secretase inhibitors that underwent human clinical trials in Alzheimer's disease and MCI patients resulted in statistically significant worsening of cognition relative to controls, which is thought to be due to its incidental effect on Notch signalling.[86]
Remove ads
Role in cardiovascular development
Summarize
Perspective
The Notch signaling pathway is a critical component of cardiovascular formation and morphogenesis in both development and disease. It is required for the selection of endothelial tip and stalk cells during sprouting angiogenesis.[87]
Cardiac development
Notch signal pathway plays a crucial role in at least three cardiac development processes: Atrioventricular canal development, myocardial development, and cardiac outflow tract (OFT) development.[88]
Atrioventricular (AV) canal development
- AV boundary formation
- Notch signaling can regulate the atrioventricular boundary formation between the AV canal and the chamber myocardium.
Studies have revealed that both loss- and gain-of-function of the Notch pathway results in defects in AV canal development.[88] In addition, the Notch target genes HEY1 and HEY2 are involved in restricting the expression of two critical developmental regulator proteins, BMP2 and Tbx2, to the AV canal.[89][90]
- AV epithelial-mesenchymal transition (EMT)
- Notch signaling is also important for the process of AV EMT, which is required for AV canal maturation. After the AV canal boundary formation, a subset of endocardial cells lining the AV canal are activated by signals emanating from the myocardium and by interendocardial signaling pathways to undergo EMT.[88] Notch1 deficiency results in defective induction of EMT. Very few migrating cells are seen and these lack mesenchymal morphology.[91] Notch may regulate this process by activating matrix metalloproteinase2 (MMP2) expression, or by inhibiting vascular endothelial (VE)-cadherin expression in the AV canal endocardium[92] while suppressing the VEGF pathway via VEGFR2.[93] In RBPJk/CBF1-targeted mutants, the heart valve development is severely disrupted, presumably because of defective endocardial maturation and signaling.[91]
Ventricular development
Some studies in Xenopus[94] and in mouse embryonic stem cells[95] indicate that cardiomyogenic commitment and differentiation require Notch signaling inhibition. Active Notch signaling is required in the ventricular endocardium for proper trabeculae development subsequent to myocardial specification by regulating BMP10, NRG1, and EphrinB2 expression.[49] Notch signaling sustains immature cardiomyocyte proliferation in mammals [96][97][98] and zebrafish.[99] A regulatory correspondence likely exists between Notch signaling and Wnt signaling, whereby upregulated Wnt expression downregulates Notch signaling, and a subsequent inhibition of ventricular cardiomyocyte proliferation results. This proliferative arrest can be rescued using Wnt inhibitors.[100]
The downstream effector of Notch signaling, HEY2, was also demonstrated to be important in regulating ventricular development by its expression in the interventricular septum and the endocardial cells of the cardiac cushions.[101] Cardiomyocyte and smooth muscle cell-specific deletion of HEY2 results in impaired cardiac contractility, malformed right ventricle, and ventricular septal defects.[102]
Ventricular outflow tract development
During development of the aortic arch and the aortic arch arteries, the Notch receptors, ligands, and target genes display a unique expression pattern.[103] When the Notch pathway was blocked, the induction of vascular smooth muscle cell marker expression failed to occur, suggesting that Notch is involved in the differentiation of cardiac neural crest cells into vascular cells during outflow tract development.
Angiogenesis
Endothelial cells use the Notch signaling pathway to coordinate cellular behaviors during the blood vessel sprouting that occurs sprouting angiogenesis.[104][105][106][107]
Activation of Notch takes place primarily in "connector" cells and cells that line patent stable blood vessels through direct interaction with the Notch ligand, Delta-like ligand 4 (Dll4), which is expressed in the endothelial tip cells.[108] VEGF signaling, which is an important factor for migration and proliferation of endothelial cells,[109] can be downregulated in cells with activated Notch signaling by lowering the levels of Vegf receptor transcript.[110] Zebrafish embryos lacking Notch signaling exhibit ectopic and persistent expression of the zebrafish ortholog of VEGF3, flt4, within all endothelial cells, while Notch activation completely represses its expression.[111]
Notch signaling may be used to control the sprouting pattern of blood vessels during angiogenesis. When cells within a patent vessel are exposed to VEGF signaling, only a restricted number of them initiate the angiogenic process. Vegf is able to induce DLL4 expression. In turn, DLL4 expressing cells down-regulate Vegf receptors in neighboring cells through activation of Notch, thereby preventing their migration into the developing sprout. Likewise, during the sprouting process itself, the migratory behavior of connector cells must be limited to retain a patent connection to the original blood vessel.[108]
Remove ads
Role in endocrine development
Summarize
Perspective
During development, definitive endoderm and ectoderm differentiates into several gastrointestinal epithelial lineages, including endocrine cells. Many studies have indicated that Notch signaling has a major role in endocrine development.
Pancreatic development
The formation of the pancreas from endoderm begins in early development. The expression of elements of the Notch signaling pathway have been found in the developing pancreas, suggesting that Notch signaling is important in pancreatic development.[112][113] Evidence suggests Notch signaling regulates the progressive recruitment of endocrine cell types from a common precursor,[114] acting through two possible mechanisms. One is the "lateral inhibition", which specifies some cells for a primary fate but others for a secondary fate among cells that have the potential to adopt the same fate. Lateral inhibition is required for many types of cell fate determination. Here, it could explain the dispersed distribution of endocrine cells within pancreatic epithelium.[115] A second mechanism is "suppressive maintenance", which explains the role of Notch signaling in pancreas differentiation. Fibroblast growth factor10 is thought to be important in this activity, but the details are unclear.[116][117]
Intestinal development
The role of Notch signaling in the regulation of gut development has been indicated in several reports. Mutations in elements of the Notch signaling pathway affect the earliest intestinal cell fate decisions during zebrafish development.[118] Transcriptional analysis and gain of function experiments revealed that Notch signaling targets Hes1 in the intestine and regulates a binary cell fate decision between adsorptive and secretory cell fates.[118]
Bone development
Early in vitro studies have found the Notch signaling pathway functions as down-regulator in osteoclastogenesis and osteoblastogenesis.[119] Notch1 is expressed in the mesenchymal condensation area and subsequently in the hypertrophic chondrocytes during chondrogenesis.[120] Overexpression of Notch signaling inhibits bone morphogenetic protein2-induced osteoblast differentiation. Overall, Notch signaling has a major role in the commitment of mesenchymal cells to the osteoblastic lineage and provides a possible therapeutic approach to bone regeneration.[53]
Remove ads
Role in cell cycle control
Summarize
Perspective
Notch signaling is critical for cell fate identity and differentiation and regulates these processes in part by controlling cell cycle progression. Specifically, Notch has been shown to promote cell cycle progression at the G1/S transition in various systems.
Photoreceptor development
In Drosophila eye development, photoreceptors undergo two waves of differentiation, where five out of eight photoreceptors differentiate in the first wave (R8, R2, R5, R3, and R4), and the other three differentiate in the second wave (R1, R6, and R7).[121] Notch has been shown to promote the second mitotic wave in Drosophila eye development.[122] Specifically, it mediates the G1/S transition by promoting dE2F activation (Drosophila E2F), a member of the E2F transcription factor family, which regulates the expression of genes important for cell proliferation, specifically those involved in the G1/S transition.[122][123] Notch does this by inhibiting RBF1 (the Drosophila homolog of the tumor suppressor Rb), which represses dE2F.[122] Additionally, Notch is required for cyclin A activation, which accumulates during the G1/S transition and may be involved in S phase onset.[122]
Health and disease
The role of Notch signaling in cell cycle regulation also has implications in health and disease. For example, Notch has been found to promote the expression of cyclin D3 and Cdk4/6 in human T-cells, thereby promoting the phosphorylation of Rb and facilitating the G1/S transition, implicating its role in cancer as several gain-of-function mutations in NOTCH1 have been identified in human acute T-cell lymphoblastic leukemias and lymphomas.[124][125] Additionally, in ventricular cardiomyocytes, which stop dividing shortly after birth, NOTCH2 signaling activation promotes cell cycle reentry.[98] It induces the expression and nuclear translocation of cyclin D, which along with Cdk4/6 promotes the phosphorylation of Rb and causes cell cycle progression through the G1/S transition.[98] This suggests that Notch signaling might regulate ventricular growth as well as cardiomyocyte regeneration, though this is unclear.[98]
Migratory identity
In the zebrafish trunk neural crest (TNC), cells migrate collectively in single-file chains, with a cell “leader” at the front of the chain that instructs the directionality of the trailing “follower” cells.[126] Notch has been found to specify cell migratory identity in the trunk neural crest – specifically, high Notch specifies leaders while low Notch specifies followers.[127] Further, cell cycle progression required for migration is regulated by Notch such that leader cells with high Notch activity quickly undergo the G1/S transition while cells with low Notch activity remain in the G1 phase for longer and thus become followers.[127]
Remove ads
Role in cancer
Summarize
Perspective
Leukemia
Aberrant Notch signaling is a driver of T cell acute lymphoblastic leukemia (T-ALL)[128] and is mutated in at least 65% of all T-ALL cases.[129] Notch signaling can be activated by mutations in Notch itself, inactivating mutations in FBXW7 (a negative regulator of Notch1), or rarely by t(7;9)(q34;q34.3) translocation. In the context of T-ALL, Notch activity cooperates with additional oncogenic lesions such as c-MYC to activate anabolic pathways such as ribosome and protein biosynthesis thereby promoting leukemia cell growth.[130]
Urothelial bladder cancer
Loss of Notch activity is a driving event in urothelial cancer. A study identified inactivating mutations in components of the Notch pathway in over 40% of examined human bladder carcinomas. In mouse models, genetic inactivation of Notch signaling results in Erk1/2 phosphorylation leading to tumorigenesis in the urinary tract.[131] As not all NOTCH receptors are equally involved in the urothelial bladder cancer, 90% of samples in one study had some level of NOTCH3 expression, suggesting that NOTCH3 plays an important role in urothelial bladder cancer. A higher level of NOTCH3 expression was observed in high-grade tumors, and a higher level of positivity was associated with a higher mortality risk. NOTCH3 was identified as an independent predictor of poor outcome. Therefore, it is suggested that NOTCH3 could be used as a marker for urothelial bladder cancer-specific mortality risk. It was also shown that NOTCH3 expression could be a prognostic immunohistochemical marker for clinical follow-up of urothelial bladder cancer patients, contributing to a more individualized approach by selecting patients to undergo control cystoscopy after a shorter time interval.[132]
Liver cancer
In hepatocellular carcinoma, for instance, it was suggesting that AXIN1 mutations would provoke Notch signaling pathway activation, fostering the cancer development, but a recent study demonstrated that such an effect cannot be detected.[133] Thus the exact role of Notch signaling in the cancer process awaits further elucidation.
Remove ads
Notch inhibitors
The involvement of Notch signaling in many cancers has led to investigation of notch inhibitors (especially gamma-secretase inhibitors) as cancer treatments which are in different phases of clinical trials.[2][134] As of 2013[update] at least 7 notch inhibitors were in clinical trials.[135] MK-0752 has given promising results in an early clinical trial for breast cancer.[136] Preclinical studies showed beneficial effects of gamma-secretase inhibitors in endometriosis,[137] a disease characterised by increased expression of notch pathway constituents.[138][139] Several notch inhibitors, including the gamma-secretase inhibitor LY3056480, are being studied for their potential ability to regenerate hair cells in the cochlea, which could lead to treatments for hearing loss and tinnitus.[140][141]
Remove ads
Mathematical modeling
Summarize
Perspective
Mathematical modeling in Notch-Delta signaling has become a pivotal tool in understanding pattern formation driven by cell-cell interactions, particularly in the context of lateral-inhibition mechanisms. The Collier model,[142] a cornerstone in this field, employs a system of coupled ordinary differential equations to describe the feedback loop between adjacent cells. The model is defined by the equations:
where and represent the levels of Notch and Delta activity in cell , respectively. Functions and are typically Hill functions, reflecting the regulatory dynamics of the signaling process. The term denotes the average level of Delta activity in the cells adjacent to cell , integrating juxtacrine signaling effects.
Recent extensions of this model incorporate long-range signaling, acknowledging the role of cell protrusions like filopodia (cytonemes) that reach non-neighboring cells.[143][144][145][146] One extended model, often referred to as the -Collier model,[143] introduces a weighting parameter to balance juxtacrine and long-range signaling. The interaction term is modified to include these protrusions, creating a more complex, non-local signaling network. This model is instrumental in exploring pattern formation robustness and biological pattern refinement, considering the stochastic nature of filopodia dynamics and intrinsic noise. The application of mathematical modeling in Notch-Delta signaling has been particularly illuminating in understanding the patterning of sensory organ precursors (SOPs) in the Drosophila's notum and wing margin.[147][148]
The mathematical modeling of Notch-Delta signaling thus provides significant insights into lateral inhibition mechanisms and pattern formation in biological systems. It enhances the understanding of cell-cell interaction variations leading to diverse tissue structures, contributing to developmental biology and offering potential therapeutic pathways in diseases related to Notch-Delta dysregulation.
Remove ads
See also
- Alagille syndrome
- Netpath – A curated resource of signal transduction pathways in humans
References
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
Remove ads









![{\displaystyle \epsilon \in [0,1]}](http://wikimedia.org/api/rest_v1/media/math/render/svg/2284041657a2d5f73d1fea4ac4af504d94382b5d)