MERS-related coronavirus
Species of virus From Wikipedia, the free encyclopedia
Middle East respiratory syndrome–related coronavirus (MERS-CoV, Betacoronavirus cameli)[1][2] or EMC/2012 (HCoV-EMC/2012), is the virus that causes Middle East respiratory syndrome (MERS).[3][4] It is a species of coronavirus which infects humans, bats, and camels.[5] The infecting virus is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the DPP4 receptor.[6] The species is a member of the genus Betacoronavirus and subgenus Merbecovirus.[7][5]
| Middle East respiratory syndrome-related coronavirus | |
|---|---|
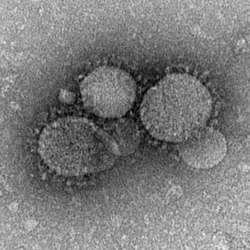 | |
| MERS-CoV particles as seen by negative stain electron microscopy. Virions contain characteristic club-like projections emanating from the viral membrane. | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
| Family: | Coronaviridae |
| Genus: | Betacoronavirus |
| Subgenus: | Merbecovirus |
| Species: | Betacoronavirus cameli |
| Synonyms | |
| |
Initially called simply novel coronavirus or nCoV, with the provisional names 2012 novel coronavirus (2012-nCoV) and human coronavirus 2012 (HCoV-12 or hCoV-12), it was first reported in June 2012 after genome sequencing of a virus isolated from sputum samples from a person who fell ill in a 2012 outbreak of a new flu-like respiratory illness. By July 2015, MERS-CoV cases had been reported in over 21 countries, in Europe, North America and Asia as well as the Middle East. MERS-CoV is one of several viruses identified by the World Health Organization (WHO) as a likely cause of a future epidemic. They list it for urgent research and development.[8][9]
Virology
Summarize
Perspective
The virus MERS-CoV is a member of the beta group of coronavirus, Betacoronavirus, lineage C. MERS-CoV genomes are phylogenetically classified into two clades, clade A and B. The earliest cases were of clade A clusters, while the majority of more recent cases are of the genetically distinct clade B.[10]
MERS-CoV is one of seven known coronaviruses to infect humans, including HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1, the original SARS-CoV (or SARS-CoV-1), and SARS-CoV-2.[11] It has frequently been referred to as a SARS-like virus.[12] By November, 2019, 2,494 cases of MERS had been reported with 858 deaths, implying a case fatality rate of greater than 30%.[13]
Early cases and spillover event
The first confirmed case was reported in Jeddah, Saudi Arabia in April 2012.[11] Egyptian virologist Ali Mohamed Zaki isolated and identified a previously unknown coronavirus from the man's lungs.[14][15][16] Zaki then posted his findings on 24 September 2012 on ProMED-mail.[15][17] The isolated cells showed cytopathic effects (CPE), in the form of rounding and syncytia formation.[17]
A second case was found in September 2012, when a 49-year-old man living in Qatar presented with similar flu symptoms. A sequence of the virus was nearly identical to that of the first case.[11]
Among animal reservoirs, CoV has a large genetic diversity yet the samples from patients suggested a similar genome, and therefore common source, though the data were limited. It was determined through molecular clock analysis that viruses from the EMC/2012 and England/Qatar/2012 date to early 2011, suggesting that these cases were descended from a single zoonotic event. It appeared the MERS-CoV had been circulating in the human population for more than a year without detection, and suggested independent transmission from an unknown source.[18][19]
Tropism

In humans, the virus has a strong tropism for nonciliated bronchial epithelial cells, and it has been shown to effectively evade the innate immune responses and antagonize interferon (IFN) production in these cells. This tropism is unique in that most respiratory viruses target ciliated cells.[20][21]
Due to the clinical similarity between MERS-CoV and SARS-CoV, it was proposed that they may use the same cellular receptor; the exopeptidase, angiotensin converting enzyme 2 (ACE2).[22] However, it was later discovered that neutralization of ACE2 by recombinant antibodies does not prevent MERS-CoV infection.[23] Further research identified dipeptidyl peptidase 4 (DPP4; also known as CD26) as a functional cellular receptor for MERS-CoV.[21] Unlike other known coronavirus receptors, the enzymatic activity of DPP4 is not required for infection. As would be expected, the amino acid sequence of DPP4 is highly conserved across species and is expressed in the human bronchial epithelium and kidneys.[21][24] Bat DPP4 genes appear to have been subject to a high degree of adaptive evolution as a response to coronavirus infections, so the lineage leading to MERS-CoV may have circulated in bat populations for a long period of time before being transmitted to people.[25]
Transmission
On 13 February 2013, the World Health Organization stated that "the risk of sustained person-to-person transmission appears to be very low."[26] The cells MERS-CoV infects in the lungs only account for 20% of respiratory epithelial cells, so a large number of virions are likely needed to be inhaled to cause infection.[24]
Anthony Fauci of the National Institutes of Health in Bethesda, Maryland, stated that MERS-CoV "does not spread in a sustained person to person way at all," while noting the possibility that the virus could mutate into a strain that does transmit from person to person.[27] However, the infection of healthcare workers has led to concerns of human to human transmission.[28]
The Centers for Disease Control and Prevention (CDC) list MERS as transmissible from human to human.[29] They state that "MERS-CoV has been shown to spread between people who are in close contact. Transmission from infected patients to healthcare personnel has also been observed. Clusters of cases in several countries are being investigated."[29][30]
However, on the 28th of May, the CDC revealed that the Illinois man who was originally thought to have been the first incidence of person-to-person spread (from the Indiana man at a business meeting), had in fact tested negative for MERS-CoV. After completing additional and more definitive tests using a neutralising antibody assay, experts at the CDC concluded that the Indiana patient did not spread the virus to the Illinois patient. Tests concluded that the Illinois man had not been previously infected. It is possible for MERS to be symptomless, and early research has shown that up to 20% of cases show no signs of active infection but have MERS-CoV antibodies in their blood.[31]
Evolution

The virus appears to have originated in bats.[32] The virus itself has been isolated from a bat.[33] This virus is closely related to the Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5.[34] Serological evidence shows that these viruses have infected camels for at least 20 years. The most recent common ancestor of several human strains has been dated to March 2012 (95% confidence interval December 2011 to June 2012).[35]
It is thought that the viruses have been present in bats for some time and had spread to camels by the mid-1990s. The viruses appear to have spread from camels to humans in the early 2010s. The original bat host species and the time of initial infection in this species has yet to be determined.[citation needed] Examination of the sequences of 238 isolates suggested that this virus has evolved into three clades differing in codon usage, host, and geographic distribution.[36]
Natural reservoir
It is believed that the virus originated in bats, one candidate being the Egyptian tomb bat.[37] Work by epidemiologist Ian Lipkin of Columbia University in New York showed that the virus isolated from a bat looked to be a match to the virus found in humans.[33][38] 2c betacoronaviruses were detected in Nycteris bats in Ghana and Pipistrellus bats in Europe that are phylogenetically related to the MERS-CoV virus.[39] However the major natural reservoir where humans get the virus infection remained unknown until on 9 August 2013, a report in the journal The Lancet Infectious Diseases showed that 50 out of 50 (100%) blood serum from Omani camels and 15 of 105 (14%) from Spanish camels had protein-specific antibodies against the MERS-CoV spike protein. Blood serum from European sheep, goats, cattle, and other camelids had no such antibodies.[40]
Soon after on 5 September 2013 a seroepidemiological study published in the journal of Eurosurveillance by R.A Perera et al.[41] where they investigated 1343 human and 625 animal sera indicated, the abundant presence of MERS-CoV specific antibody in 108 out of 110 Egyptian dromedary camels but not in other animals such as goats, cows or sheep in this region.[41] These are the first and significant scientific reports that indicated the role of "dromedary camels" as a reservoir of MERS-CoV.[citation needed]
Research has linked camels, showing that the coronavirus infection in dromedary camel calves and adults is a 99.9% match to the genomes of human clade B MERS-CoV.[42] At least one person who has fallen sick with MERS was known to have come into contact with camels or recently drank camel milk.[43] Countries like Saudi Arabia and the United Arab Emirates produce and consume large amounts of camel meat. The possibility exists that African or Australian bats harbor the virus and transmit it to camels. Imported camels from these regions might have carried the virus to the Middle East.[44]
In 2013 MERS-CoV was identified in three members of a dromedary camel herd held in a Qatar barn, which was linked to two confirmed human cases who have since recovered. The presence of MERS-CoV in the camels was confirmed by the National Institute of Public Health and Environment (RIVM) of the Ministry of Health and the Erasmus Medical Center (WHO Collaborating Center), the Netherlands. None of the camels showed any sign of disease when the samples were collected. The Qatar Supreme Council of Health advised in November 2013 that people with underlying health conditions, such as heart disease, diabetes, kidney disease, respiratory disease, the immunosuppressed, and the elderly, avoid any close animal contacts when visiting farms and markets, and to practice good hygiene, such as washing hands.[45]
A further study on dromedary camels from Saudi Arabia published in December 2013 revealed the presence of MERS-CoV in 90% of the evaluated dromedary camels (310), suggesting that dromedary camels not only could be the main reservoir of MERS-CoV, but also the animal source of MERS.[46]
According to the 27 March 2014 MERS-CoV summary update, recent studies support that camels serve as the primary source of the MERS-CoV infecting humans, while bats may be the ultimate reservoir of the virus. Evidence includes the frequency with which the virus has been found in camels to which human cases have been exposed, seriological data which shows widespread transmission in camels, and the similarity of the camel CoV to the human CoV.[47]
On 6 June 2014, the Arab News newspaper highlighted the latest research findings in the New England Journal of Medicine in which a 44-year-old Saudi man who kept a herd of nine camels died of MERS in November 2013. His friends said they witnessed him applying a topical medicine to the nose of one of his ill camels—four of them reportedly sick with nasal discharge—seven days before he himself became stricken with MERS. Researchers sequenced the virus found in one of the sick camels and the virus that killed the man, and found that their genomes were identical. In that same article, the Arab News reported that as of 6 June 2014, there have been 689 cases of MERS reported within the Kingdom of Saudi Arabia with 283 deaths.[48]
Taxonomy
MERS-CoV is more closely related to the bat coronaviruses HKU4 and HKU5 (lineage 2C) than it is to SARS-CoV (lineage 2B) (2, 9), sharing more than 90% sequence identity with their closest relationships, bat coronaviruses HKU4 and HKU5 and therefore considered to belong to the same species by the International Committee on Taxonomy of Viruses (ICTV).[citation needed]
- Scientific name: Betacoronavirus cameli[1]
- Common name: Middle East respiratory syndrome coronavirus ( MERS-CoV) [2]
- Other names:
- novel coronavirus (nCoV)
- London1 novel CoV/2012[49]
- Human Coronavirus Erasmus Medical Center/2012 (HCoV-EMC/2012)
Research and patent
Summarize
Perspective
Saudi officials had not given permission for Dr. Zaki, the first isolator of the human strain, to send a sample of the virus to Fouchier and were angered when Fouchier claimed the patent on the full genetic sequence of MERS-CoV.[50]
The editor of The Economist observed, "Concern over security must not slow urgent work. Studying a deadly virus is risky. Not studying it is riskier."[50] Dr. Zaki was fired from his job at the hospital as a result of bypassing the Saudi Ministry of Health in his announcement and sharing his sample and findings.[51][52][53][54]
At their annual meeting of the World Health Assembly in May 2013, WHO chief Margaret Chan declared that intellectual property, or patents on strains of new virus, should not impede nations from protecting their citizens by limiting scientific investigations. Deputy Health Minister Ziad Memish raised concerns that scientists who held the patent for MERS-CoV would not allow other scientists to use patented material and were therefore delaying the development of diagnostic tests.[55] Erasmus MC responded that the patent application did not restrict public health research into MERS and MERS-CoV,[56] and that the virus and diagnostic tests were shipped—free of charge—to all that requested such reagents.
Mapping
There are a number of mapping efforts focused on tracking MERS coronavirus. On 2 May 2014, the Corona Map[57] was launched to track the MERS coronavirus in realtime on the world map. The data is officially reported by WHO or the Ministry of Health of the respective country.[58] HealthMap also tracks case reports with inclusion of news and social media as data sources as part of HealthMap MERS.[59]
See also
References
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
