Methionine synthase
Mammalian protein found in Homo sapiens From Wikipedia, the free encyclopedia
Methionine synthase (MS, MeSe, MTR) is primarily responsible for the regeneration of methionine from homocysteine in most individuals. In humans it is encoded by the MTR gene (5-methyltetrahydrofolate-homocysteine methyltransferase).[5][6] Methionine synthase forms part of the S-adenosylmethionine (SAMe) biosynthesis and regeneration cycle,[7] and is the enzyme responsible for linking the cycle to one-carbon metabolism via the folate cycle. There are two primary forms of this enzyme, the Vitamin B12 (cobalamin)-dependent (MetH) and independent (MetE) forms,[8] although minimal core methionine synthases that do not fit cleanly into either category have also been described in some anaerobic bacteria.[9] The two dominant forms of the enzymes appear to be evolutionary independent and rely on considerably different chemical mechanisms.[10] Mammals and other higher eukaryotes express only the cobalamin-dependent form. In contrast, the distribution of the two forms in Archaeplastida (plants and algae) is more complex. Plants exclusively possess the cobalamin-independent form,[11] while algae have either one of the two, depending on species.[12] Many different microorganisms express both the cobalamin-dependent and cobalamin-independent forms.[13]
| MTR | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | MTR, HMAG, MS, cblG, 5-methyltetrahydrofolate-homocysteine methyltransferase | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 156570; MGI: 894292; HomoloGene: 37280; GeneCards: MTR; OMA:MTR - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| EC number | 2.1.1.13 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Mechanism
Summarize
Perspective

Methionine synthase catalyzes the final step in the regeneration of methionine (Met) from homocysteine (Hcy). Both the cobalamin-dependent and cobalamin-independent forms of the enzyme carry out the same overall chemical reaction, the transfer of a methyl group from 5-methyltetrahydrofolate (N5-MeTHF) to homocysteine, yielding tetrahydrofolate (THF) and methionine.[8] Methionine synthase is the only mammalian enzyme that metabolizes N5-MeTHF to regenerate the active cofactor THF. In the cobalamin-dependent (MetH) form of the enzyme, the reaction proceeds by two steps in a preferred ordered sequential mechanism.[14] The physiological resting state of the enzyme is thought to contain the enzyme-bound(Cob) cofactor in the methylcobalamin form, with the cobalt atom in the formal +3 valence state (Cob(III)-Me). The cobalamin is then demethylated by zinc-activated thiolate homocysteine, generating methionine and reducing the cofactor to a Cob(I) state. When in the Cob(I) form, the enzyme-bound cofactor is now able to abstract a methyl group from activated 5-methyltetrahydrofolate (N5-MeTHF), yielding tetrahydrofolate (THF) and regenerating the methylcoalamin form of the enzyme.[15]

Under physiological conditions, approximately once every 2000 catalytic turnovers the Co(I) may be oxidized into inactive Co(II) in cob-dependent MetH. To account for this effect, the protein contains a self-reactivation mechanism, a reductive methylation process that uses S-adenosylmethionine as a distinct methyl donor. In humans, the enzyme is reduced in this process by methionine synthase reductase (MTRR), which consists of flavodoxin-like and ferrodoxin-NADP+ oxidoreductase (FNR)-like domains.[16] In many bacteria, the reduction is carried out by a single domain flavodoxin protein.[17] The reductase protein is responsible for transfer of an electron from a reduced FMN cofactor to the inactive Cob(II), which enables regeneration of the active methylcobalamin enzyme via methyl transfer from S-adenosylmethionine to the reduced Cob(I) intermediate.[18] This process is known as the reactivation cycle, and is thought to be gated from the normal catalytic cycle by large-scale conformational rearrangements within the enzyme.[19] Because the oxidation of Cob(I) inevitably shuts down cob-dependent methionine synthase activity, defects or deficiencies in methionine synthase reductase have been implicated in some of the disease associations for methionine synthase deficiency.[20]
The mechanism of the cobalamin-independent (MetE) form, by contrast, proceeds through a direct methyl transfer from the activated N5-MeTHF to zinc thiolate homocysteine. Although the mechanism is considerably simpler, the direct transfer reaction is much less favorable than the cobalamin-mediated reactions and as a result the turnover rate for MetE is ~100x slower than that of MetH. As it does not contain the cobalamin cofactor, the cobalamin-independent enzyme is not prone to oxidative inactivation [21][8][22][23]
Structure
Summarize
Perspective

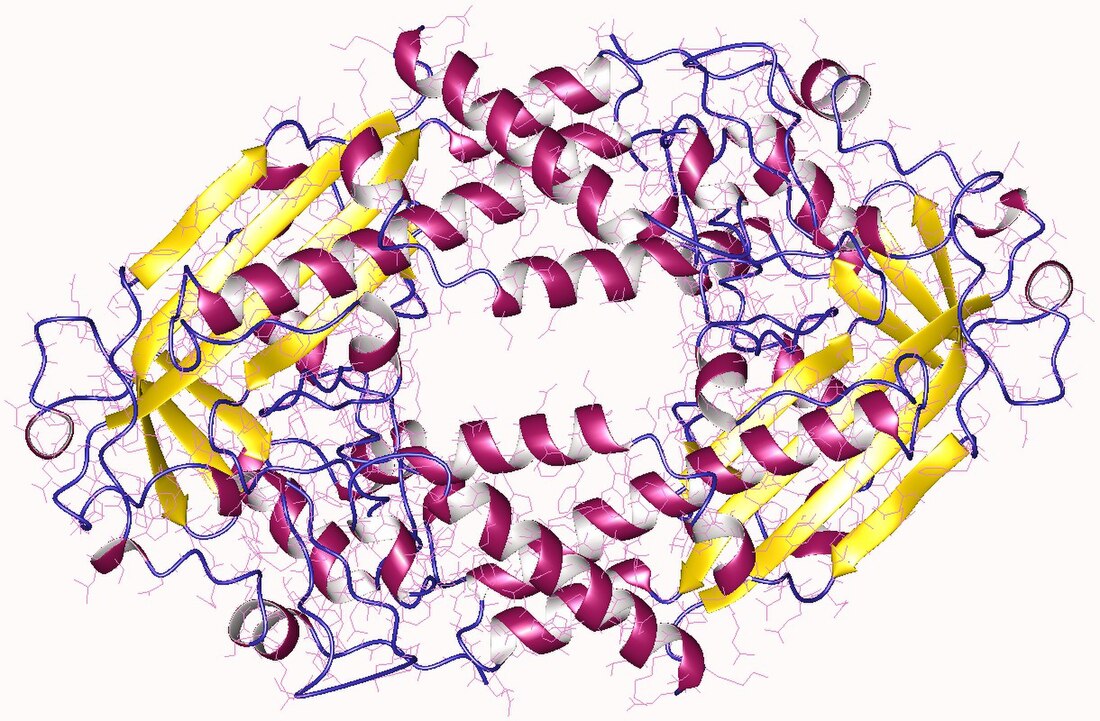
High-resolution structures have been solved by X-ray crystallography for intact MetE both in the absence and presence of substrates[23][22] and for fragments of MetH,[24][25][26][27] although no structural description exists of a fully intact MetH enzyme. The available structures and accompanying bioinformatic analysis indicate minimal similarity in the overall structure, although there are similarities within the substrate-binding sites themselves.[28] Cob-dependent MetH is divided into 4 separate domains. The domains, from N- to C-terminus, are denoted homocysteine binding (Hcy domain), N5-methylTHF binding (MTHF domain) Cobalamin-binding (Cob domain) and the S-adenosymethionine-binding or reactivation domain. The reactivation domain binds SAM and is the site of interaction with flavodoxin or Methionine Synthase Reductase during the reactivation cycle of the enzyme.[17][16][20] The cobalamin-binding domain contains two subdomains, with the cofactor bound to the Rossman-fold B12-binding subdomain, which is in turn capped by the other subdomain, the four-helix bundle cap subdomain.[25] The four-helix bundle serves to protect the cobalamin cofactor from unwanted reactivity, but can significantly change conformations to expose the cofactor allow it access to the other substrates during turnover.[26] Both the Hcy and N5-MeTHF domains adopt a TIM barrel architecture; the Hcy domain contains the zinc-binding site, which in MetH consists of three cysteine residues coordinated to a zinc ion which in turn binds and activates Hcy. The N5-MeTHF binding domain binds and activates N5-MeTHF via a hydrogen bonding network with several asparagine, arginine, and aspartic acid residues. During turnover, the enzyme undergoes significant conformational changes that involve moving the Cob-domain back and forth from the Hcy domain to the N5-MeTHF domain in order for the two methyl transfer reactions to proceed.[24]
The cob-independent MetE consists of two TIM-barrel domains that bind homocysteine and N5-MeTHF individually. The two domains adopt a face-to-face double barrel architecture, which requires a "closing" of the structure upon binding of both substrates to enable the direct methyl transfer.[22] Substrate-binding strategies are similar to MetH, although in the case of MetE the zinc atom is instead coordinated to two cysteines, a histidine and a glutamate,[23] for which an example is shown on the right.
Biochemical function
Summarize
Perspective

In humans the enzyme's main purpose is to regenerate Met in the S-adenosylmethionine (SAM) cycle. The SAM cycle in a single turnover consumes Met and ATP and generates Hcy, and can involve any of a number of critical enzymatic reactions that use S-adenosylmethionine as the source of an active methyl group for methylation of nucleic acids, histones, phospholipids and various proteins.[29][30] As such, methionine synthase serves an essential function by allowing the SAM cycle to perpetuate without a constant influx of Met. As a secondary effect, methionine synthase also serves to maintain low levels of Hcy and, because methionine synthase is one of the few enzymes that used N5-MeTHF as a substrate, to indirectly maintain THF levels.[31][32]
In bacteria and plants, methionine synthase serves a dual purpose of both perpetuating the SAM cycle and catalyzing the final synthetic step in the de novo synthesis of Met, which is one of the 20 canonical amino acids.[33][11] While the chemical reaction is exactly the same for both processes, the overall function is distinct from methionine synthase in humans because Met is an essential amino acid that is not synthesized de novo in the body.[34]
Clinical significance
Mutations in the MTR gene have been identified as the underlying cause of methylcobalamin deficiency complementation group G, or methylcobalamin deficiency cblG-type.[5] Deficiency or deregulation of the enzyme due to deficient methionine synthase reductase can directly result in elevated levels of homocysteine (hyperhomocysteinemia), which is associated with blindness, neurological symptoms, and birth defects.[35][36] Methionine synthase reductase (MTRR) or methylene-tetrahydrofolate reductase (MTHFR) deficiencies can also result in the condition. Most cases of methionine synthase deficiency are symptomatic within 2 years of birth with many patients rapidly developing severe encephalopathy.[37] One consequence of reduced methionine synthase activity that is measurable by routine clinical blood tests is megaloblastic anemia.
Genetics
Several cblG-associated polymorphisms in the MTR gene have been identified.[38]
- 2756D→G (Asp919Gly)
- 3804C→T (Pro1137Leu)
- Δ2926A-2928T (ΔIle881)
See also
References
Further reading
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.