Loading AI tools
Antibody fragment From Wikipedia, the free encyclopedia
A single-domain antibody (sdAb), also known as a Nanobody, is an antibody fragment consisting of a single monomeric variable antibody domain. Like a whole antibody, it is able to bind selectively to a specific antigen. With a molecular weight of only 12–15 kDa, single-domain antibodies are much smaller than common antibodies (150–160 kDa) which are composed of two heavy protein chains and two light chains, and even smaller than Fab fragments (~50 kDa, one light chain and half a heavy chain) and single-chain variable fragments (~25 kDa, two variable domains, one from a light and one from a heavy chain).[1]

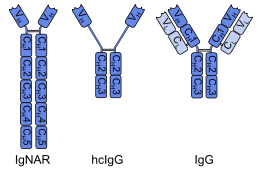
The first single-domain antibodies were engineered from heavy-chain antibodies found in camelids; these are called VHH fragments. Cartilaginous fishes also have heavy-chain antibodies (IgNAR, 'immunoglobulin new antigen receptor'), from which single-domain antibodies called VNAR fragments can be obtained.[2] An alternative approach is to split the dimeric variable domains from common immunoglobulin G (IgG) from humans or mice into monomers. Although most research into single-domain antibodies is currently based on heavy chain variable domains, Nanobodies derived from light chains have also been shown to bind specifically to target epitopes.[3]
Camelid Nanobodies have been shown to be just as specific as antibodies, and in some cases they are more robust. They are easily isolated using the same phage panning procedure used for antibodies, allowing them to be cultured in vitro in large concentrations. The smaller size and single domain make these antibodies easier to transform into bacterial cells for bulk production, making them ideal for research purposes.[4]
Single-domain antibodies are being researched for multiple pharmaceutical applications, and have potential for use in the treatment of acute coronary syndrome, cancer, Alzheimer's disease,[5][6] and Covid-19.[7][8][9]
A single-domain antibody is a peptide chain of about 110 amino acids long, comprising one variable domain (VH) of a heavy-chain antibody, or of a common IgG. These peptides have similar affinity to antigens as whole antibodies, but are more heat-resistant and stable towards detergents and high concentrations of urea. Those derived from camelid and fish antibodies are less lipophilic and more soluble in water, owing to their complementarity-determining region 3 (CDR3), which forms an extended loop (coloured orange in the ribbon diagram above) covering the lipophilic site that normally binds to a light chain.[10][11] In contrast to common antibodies, two out of six single-domain antibodies survived a temperature of 90 °C (194 °F) without losing their ability to bind antigens in a 1999 study.[12] Stability towards gastric acid and proteases depends on the amino acid sequence. Some species have been shown to be active in the intestine after oral application,[13][14] but their low absorption from the gut impedes the development of systemically active orally administered single-domain antibodies.

The comparatively low molecular mass leads to a better permeability in tissues, and to a short plasma half-life since they are eliminated renally.[1] Unlike whole antibodies, they do not show complement system triggered cytotoxicity because they lack an Fc region. Camelid and fish derived sdAbs are able to bind to hidden antigens that are not accessible to whole antibodies, for example to the active sites of enzymes.[15] This property has been shown to result from their extended CDR3 loop, which is able to penetrate such buried sites.[11][16][15]
A single-domain antibody can be obtained by immunization of dromedaries, camels, llamas, alpacas or sharks with the desired antigen and subsequent isolation of the mRNA coding for the variable region (VNAR and VHH) of heavy-chain antibodies. Large phage displayed VNAR and VHH single domain libraries were established from nurse sharks[17] and dromedary camels.[18][19] Screening techniques like phage display and ribosome display help to identify the clones binding the antigen.[20][17][21][18][22][8][19] The single domain antibodies including VNARs can be humanized for clinical applications.[23]
Alternatively, single-domain antibodies can be made from common murine,[24] rabbit[25] or human IgG[26] with four chains.[27] The process is similar, comprising gene libraries from immunized or naïve donors and display techniques for identification of the most specific antigens. A problem with this approach is that the binding region of common IgG consists of two domains (VH and VL), which tend to dimerize or aggregate because of their lipophilicity. Monomerization is usually accomplished by replacing lipophilic by hydrophilic amino acids, but often results in a loss of affinity to the antigen.[28] If affinity can be retained, the single-domain antibodies can likewise be produced in E. coli,[25][26][29] S. cerevisiae or other organisms.
Humans occasionally produce single domain antibodies by the random creation of a stop codon in the light chain. Human single-domain antibodies targeting various tumor antigens including mesothelin,[29] GPC2[30] and GPC3[26][31] were isolated by phage display. The HN3 human single-domain antibodies have been used to create immunotoxins [31][32][33] and chimeric antigen receptor (CAR) T cells[34] for treating liver cancer. Blocking the Wnt binding domain of GPC3 by the HN3 human single-domain antibody inhibits Wnt activation in liver cancer cells.[35]
Single-domain antibodies allow a broad range of applications in biotechnical as well as therapeutic use due to their small size, simple production and high affinity.[36][37][15]
The fusion of a fluorescent protein to a Nanobody generates a so-called chromobody. Chromobodies can be used to recognize and trace targets in different compartments of living cells. They can therefore increase the possibilities of live cell microscopy and will enable novel functional studies.[38] The coupling of an anti-GFP Nanobody to a monovalent matrix, called GFP-nanotrap, allows the isolation of GFP-fusion proteins and their interacting partners for further biochemical analyses.[39] Single molecule localization with super-resolution imaging techniques requires the specific delivery of fluorophores into close proximity with a target protein. Due to their large size the use of antibodies coupled to organic dyes can often lead to a misleading signal owing to the distance between the fluorophore and the target protein. The fusion of organic dyes to anti-GFP Nanobodies targeting GFP-tagged proteins allows nanometer spatial resolution and minimal linkage error because of the small size and high affinity.[40] The size dividend of Nanobodies also benefits the correlative light-electron microscopy study. Without any permeabilization agent, the cytoplasm of the chemically fixed cells are readily accessible to the fluorophore tagged Nanobodies. Their small size also allows them to penetrate deeper into volumetric samples than regular antibodies. High ultrastructural quality is preserved in the tissue that is imaged by fluorescence microscope and then electron microscope. This is especially useful for the neuroscience research that requires both molecular labeling and electron microscopic imaging.[41]
In diagnostic biosensor applications Nanobodies may be used prospectively as a tool. Due to their small size, they can be coupled more densely on biosensor surfaces. In addition to their advantage in targeting less accessible epitopes, their conformational stability also leads to higher resistance to surface regeneration conditions. After immobilizing single-domain antibodies on sensor surfaces sensing human prostate-specific antigen (hPSA) were tested. The Nanobodies outperformed the classical antibodies in detecting clinical significant concentrations of hPSA.[42]
To increase the crystallization probability of a target molecule, Nanobodies can be used as crystallization chaperones. As auxiliary proteins, they can reduce the conformational heterogeneity by binding and stabilizing just a subset of conformational states. They also can mask surfaces interfering with the crystallization while extending regions that form crystal contacts.[43][37]

Single-domain antibodies have been tested as a new therapeutic tool against multiple targets. In mice infected with influenza A virus subtype H5N1, Nanobodies directed against hemaglutinin suppressed replication of the H5N1 virus in vivo and reduced morbidity and mortality.[44] Nanobodies targeting the cell receptor binding domain of the virulence factors toxin A and toxin B of Clostridioides difficile were shown to neutralize cytopathic effects in fibroblasts in vitro.[45] Nanobody conjugates recognizing antigen presenting cells have been successfully used for tumor detection[46] or targeted antigen delivery to generate strong immune response.[47]
Orally available single-domain antibodies against E. coli-induced diarrhoea in piglets have been developed and successfully tested.[14] Other diseases of the gastrointestinal tract, such as inflammatory bowel disease and colon cancer, are also possible targets for orally available single-domain antibodies.[48]
Detergent-stable species targeting a surface protein of Malassezia furfur have been engineered for use in anti-dandruff shampoos.[10]
As an approach for photothermal therapy Nanobodies binding to the HER2 antigen, which is overexpressed in breast and ovarian cancer cells, were conjugated to branched gold nanoparticles (see figure). Tumor cells were destroyed photothermally using a laser in a test environment.[49]
Caplacizumab, a single-domain antibody targeting von Willebrand factor is in clinical trials for the prevention of thrombosis in patients with acute coronary syndrome.[50] A Phase II study examining ALX-0081 in high risk percutaneous coronary intervention has started in September 2009.[51]
Ablynx expects that their Nanobodies might cross the blood–brain barrier and permeate into large solid tumours more easily than whole antibodies, which would allow for the development of drugs against brain cancers.[48]
Nanobodies that tightly bind to the RBD domain of the spike protein of betacoronaviruses (including SARS-CoV-2 which causes COVID-19) and blocks interactions of spike with the cell receptor ACE2, has been recently identified[52][18]
Application of various single domain antibodies (Nanobodies) for the prevention and treatment of infection by various highly pathogenic human coronaviruses (HPhCoVs) has been reported. The prospects, potency and challenges of deploying Nanobodies to bind and neutralize SARS-CoV-2 and akin have been recently highlighted.[53]
One of the most common causes of nagana – Trypanosoma brucei brucei – can be targeted by sdAbs. Stijlemans et al. 2004 succeeded in inducing effective sdAbs from rabbit and Camelus dromedarius by displaying a variable surface glycoprotein antigen to the vertebrates' immune systems using a phage. In the future, these therapies will surpass natural antibodies by reaching locations currently unreachable due to natural antibodies' larger size.[54]
Seamless Wikipedia browsing. On steroids.
Every time you click a link to Wikipedia, Wiktionary or Wikiquote in your browser's search results, it will show the modern Wikiwand interface.
Wikiwand extension is a five stars, simple, with minimum permission required to keep your browsing private, safe and transparent.